Time-resolved fluorescence anisotropy analysis.
Project description
Data analysis of time-resolved fluorescence anisotropy measurements (TRAMs).
TRAMs are state-of-the-art techniques that can be used to analyse protein function and interaction.
Why use TRAM techniques?
To measure molecule rotation speeds (e.g. protein size, structure, ligand binding).
To measure oligomerisation properties, via Foerster Resonance Energy Transfer (FRET) between two fluorescent molecules.
What is blitzcurve for?
fitting curves to experimental TRAM data
extracting useful fit parameters
comparing samples
How does the experiment work?
excitation of fluorescent molecules
measurement of the depolorisation of the emitted light (polarisation / anisotropy)
time-resolved methods: pulse excitation, and measurement of the change in anisotropy over time in nanoseconds
Analysis methods
appropriate fitting methods are still under development
current input: csv with anisotropy and time (ns) values
- current fitting methods for anisotropy vs time
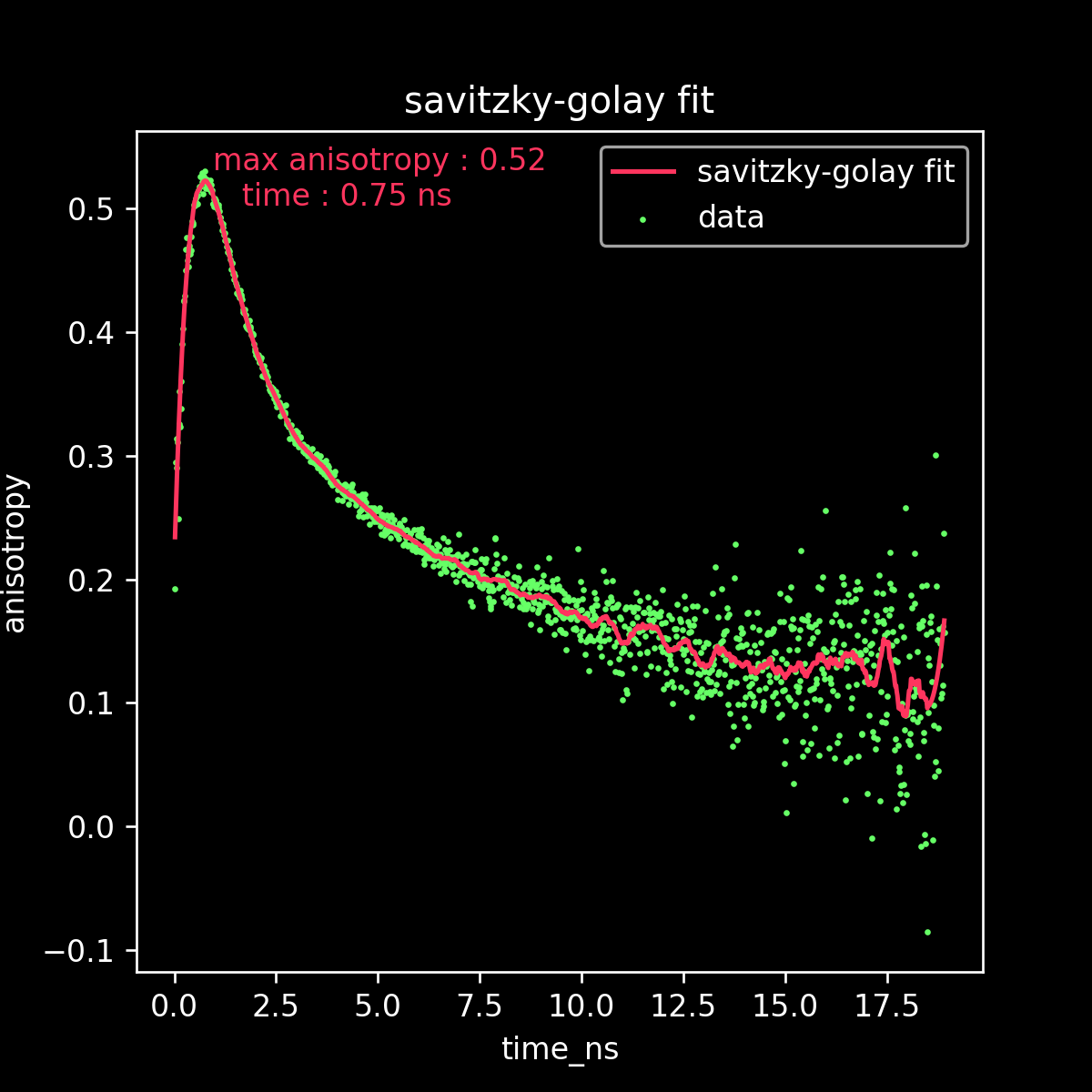
Savitzky Golay fit to all raw data
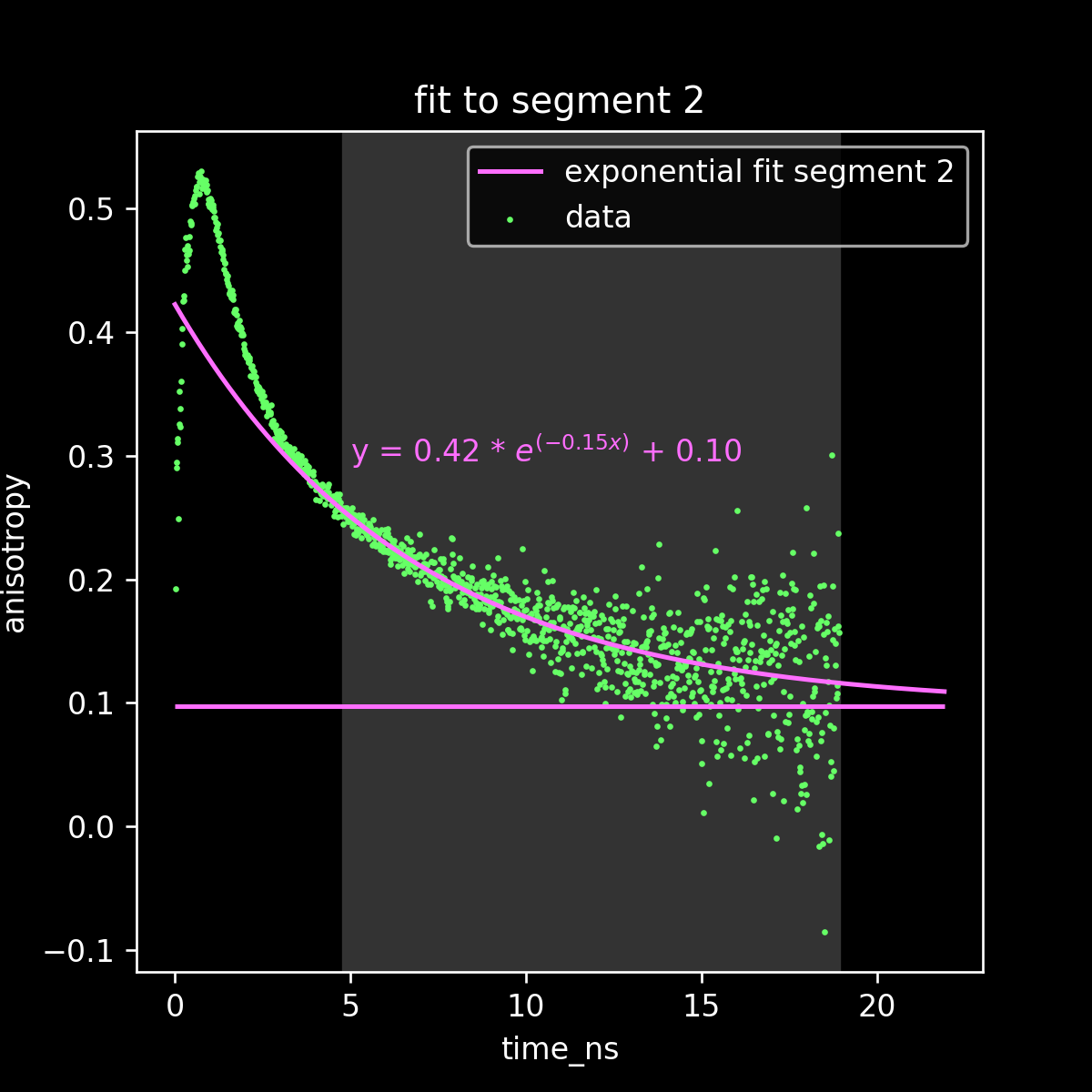
Exponential fit to initial decay data
Exponential fit to final decay data
- key measured parameters:
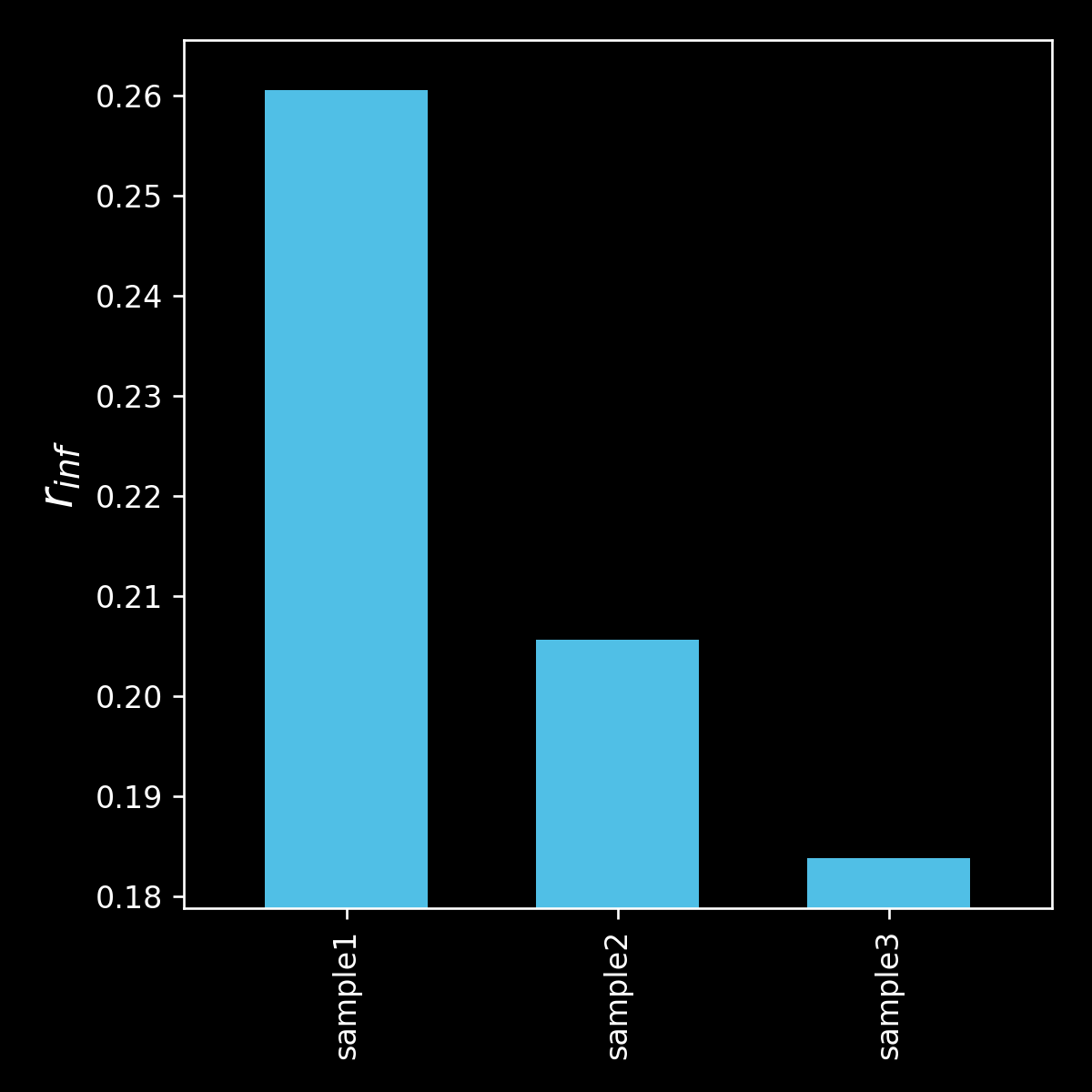
r_inf (predicted anisotropy at an infinite range in time)
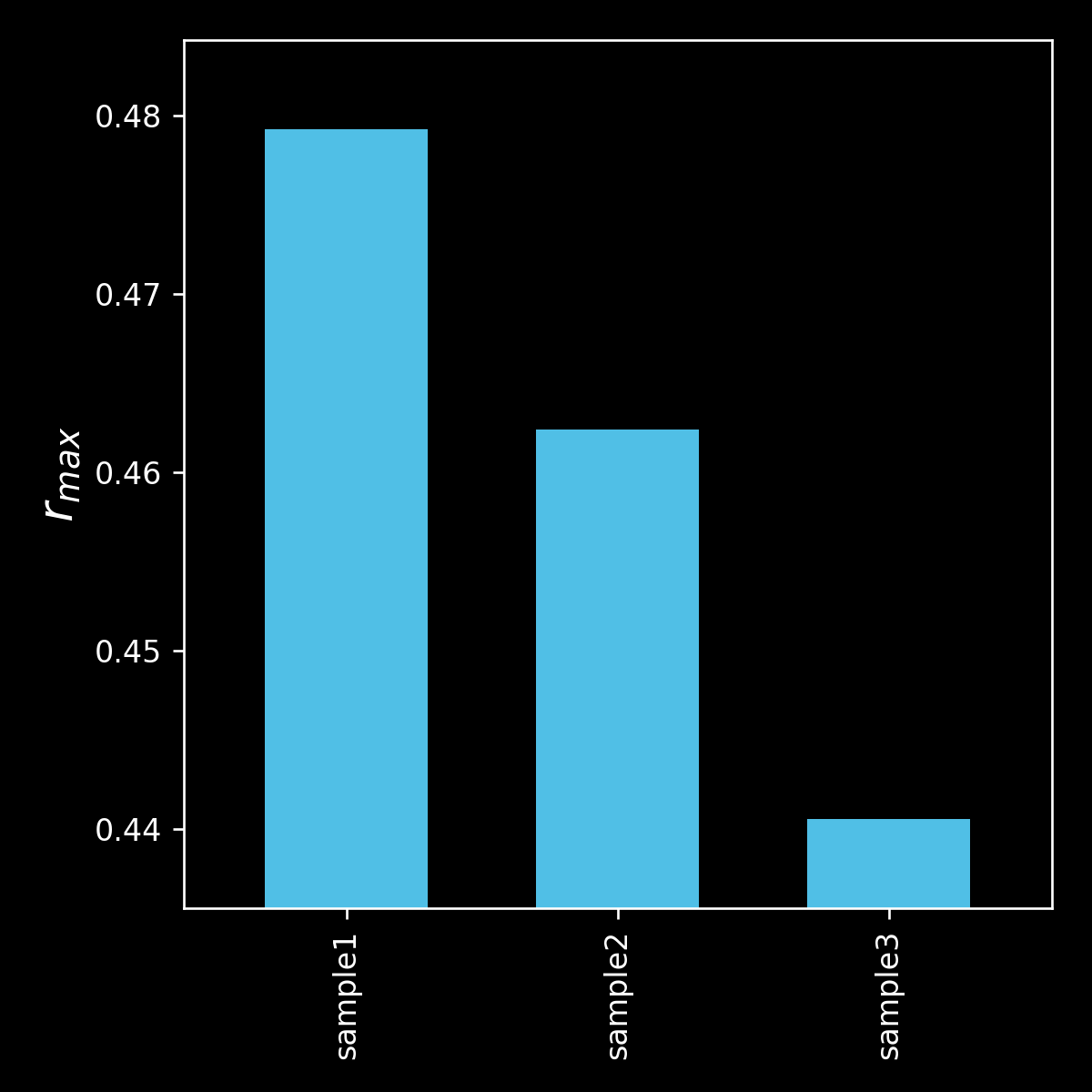
r_max (maximum anisotropy measured at any timepoint)
Installation
pip install blitzcurve
Blitzcurve should be compatable with Anaconda python 3.x or any scientific python package
Usage
import blitzcurve
# define data directory with csv files
data_dir = r"D:\data\20180229_TRdata"
# OPTIONAL: define which data files will be analysed
file_list = ["10nM-FGC1-2min_aniso.txt", "10nM-FGC2-2min_aniso.txt"]
# run blitzcurve function to fit curves to individual samples
blitzcurve.run_fit(data_dir, figs_to_plot=file_list)
# setup a dictionary to shorten long sample names
name_dict = {"10nM-FGC1-2min_aniso.txt": "FGC1", "10nM-FGC2-2min_aniso.txt": "FGC2", "10nM-FGC3-2min_aniso.txt": "FGC3"}
# run blitzcurve function to compare curves and parameters for multiple samples
blitzcurve.run_compare(data_dir, name_dict=name_dict)Contribute
Collaborators and pull requests are welcome. Send us an email.
License
This python package is released under the permissive MIT license.
Contact
Contact details are available at the staff pages of Mark Teese or Philipp Heckmeier within the Langosch lab of the Technical University of Munich.

Examples
fit to obtain r_max
fit to obtain r_inf
barchart comparing r_max
barchart comparing r_inf
linechart comparing fit to full data for three samples
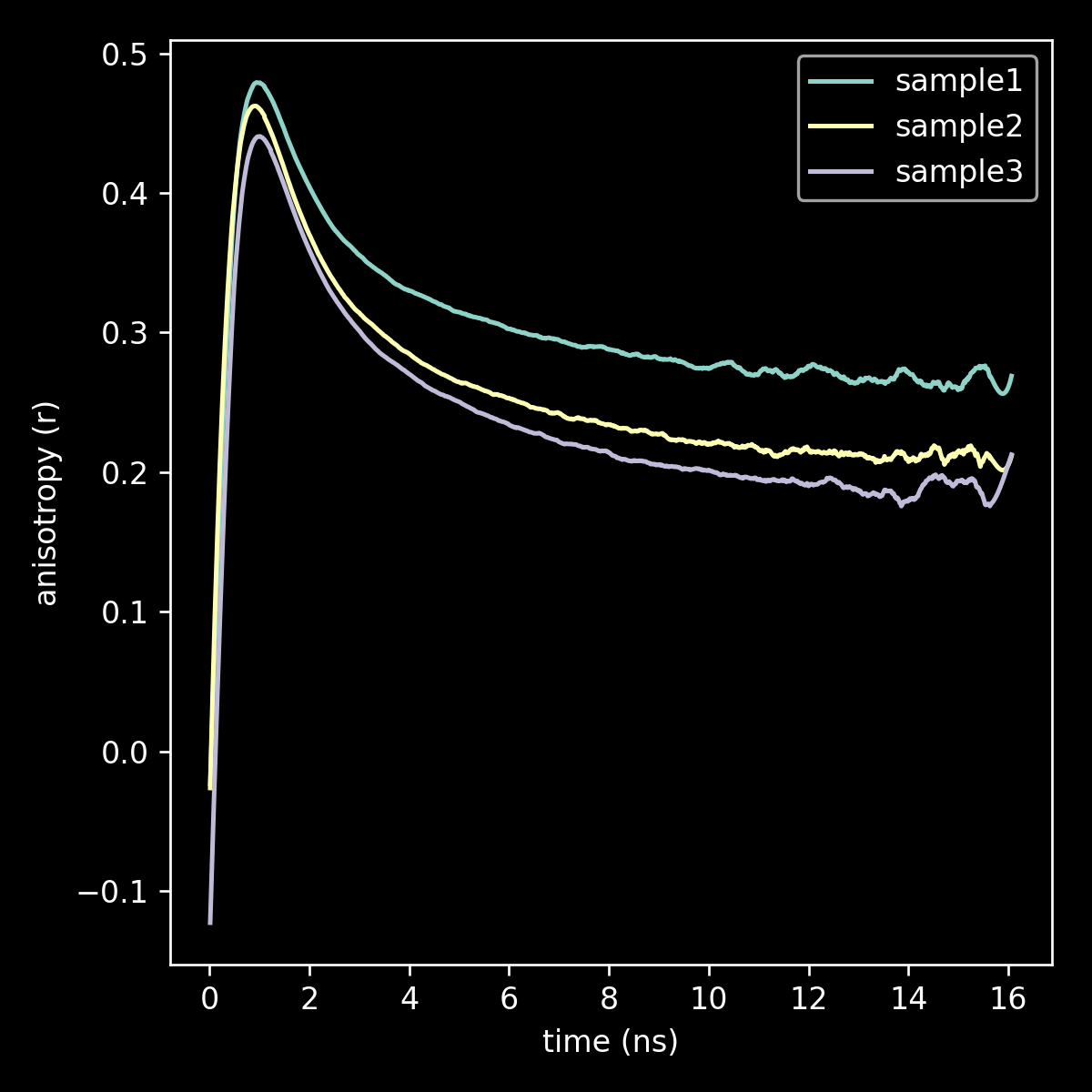
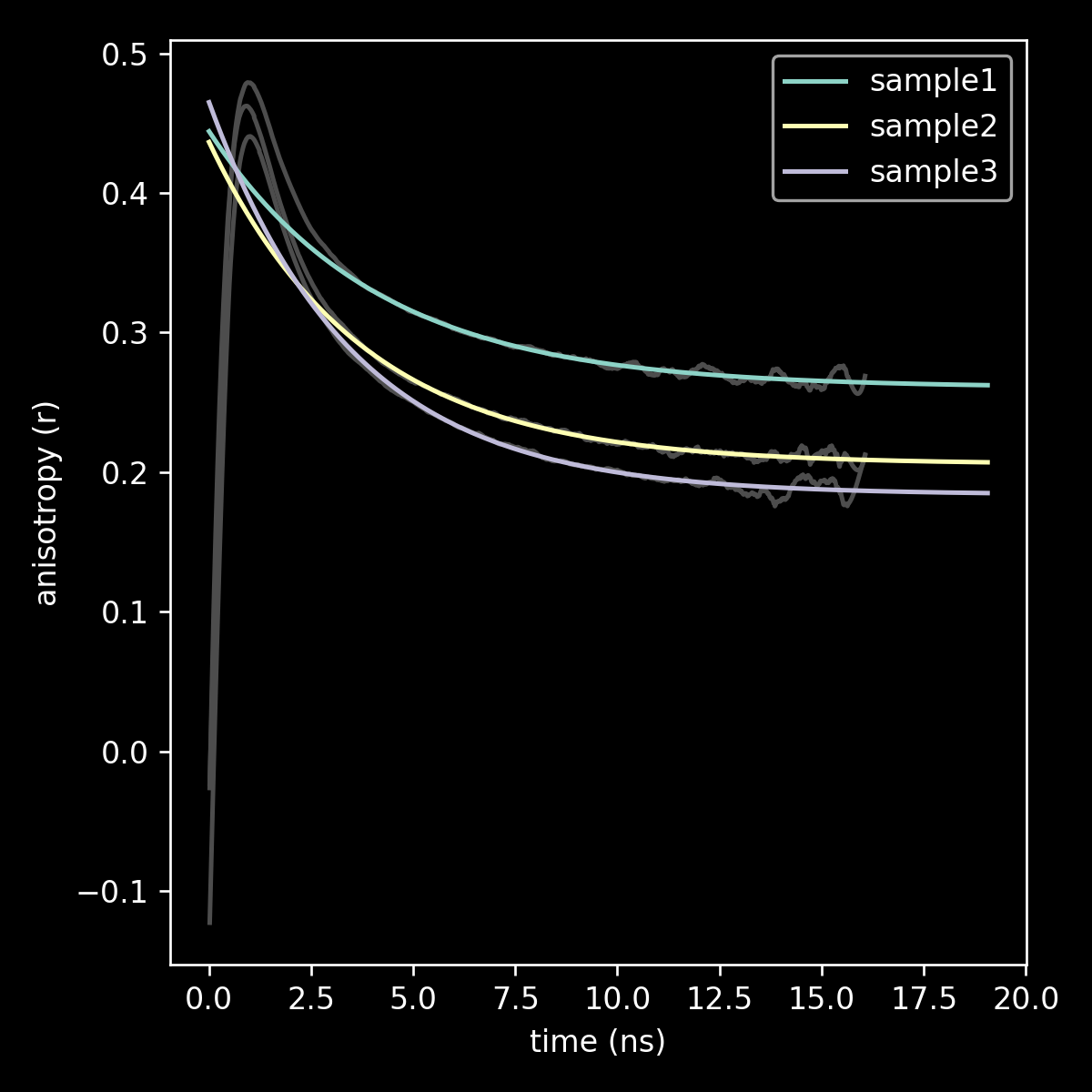
linechart comparing fit to r_inf for three samples
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Hashes for blitzcurve-0.0.2-py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 66f6beafde91ce7e7dec15a69dc09088e086b061f23ffe51a30979d9ad1c5ec6 |
|
| MD5 | 8d2ba40efba330577e2abfb173d2a9fe |
|
| BLAKE2b-256 | dc10e45088ce2f282157d822d9f41df33d8daaba078fe2b23e07fe097277ec49 |










