Python implementation of the kinetic model of neuromuscular transmission dynamics.
Project description
Dynamics of Neuromuscular Transmission Reproduced by Calcium-Dependent and Reversible Serial Transitions in the Vesicle Fusion Complex
KiNeuron: Python implementation of the kinetic model of neuromuscular transmission dynamics.
KiNeuron is an open-source implementation of our mechanistic kinetic model of neuromuscular transmission based on sequential maturation transitions in the molecular fusion complex.
KiNeuron is:
- Simple -- It is possible to simulate an alternative kinetic model with a few lines of code. This way, you can focus on the key parts of the problem that really matter.
- Flexible -- It is possible to customise the kinetic model by adjusting the number of transition states, the kinetic transitions between states, as well as the rate constants. Further, KiNeuron allows the addition of a stimulation protocol.
- Source Code: https://github.com/alexini-mv/kinetic-neurotransmission
- Bug reports: https://github.com/alexini-mv/kinetic-neurotransmission/issues
- Citing in your work: https://doi.org/10.3389/fnsyn.2021.785361
Requirements
KiNeuron requires:
- python >= 3.6
- graphviz >= 0.19.1
- matplotlib >= 3.3.4
- numpy >= 1.19.5
- pandas >= 1.1.5
- tqdm >= 4.62.3
To use the graph display functions of the model, it is necessary to install Graphviz backend for your OS as described in following documentation.
Installation
There are two ways to install KiNeuron:
-
Via PyPI repository (recommended):
- In your local workspace, create a new Python Virtual Environment with venv or conda.
- Install KiNeuron as follow:
$ python -m pip install -U kineuron
- All dependencies are downloaded and installed. To verify that it has been installed correctly, you can check the following output:
$ python -c "import kineuron; print(kineuron.__version__)" '0.1.3'
-
Via GitHub:
- Clone this project repository to your local workspace.
- Create a new Python Virtual Environment with venv or conda.
- Install the required libraries from the file
requeriments.txtinto your virtual environment as follow:
$ python -m pip install -r requeriments.txt
You are ready to use KiNeuron.
Usage Example
Creating a Model
Create a file main.py and import the following classes:
from kineuron import (KineticModel, RateConstant, Solver, Stimulation,
Transition, TransitionState)
Then, instantiate the model objects as follows:
model = KineticModel(name='my-model', vesicles=100)
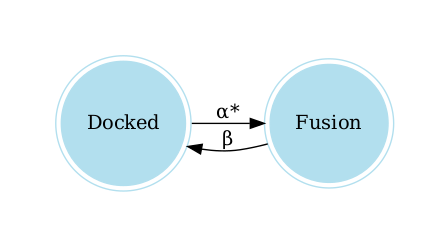
docked = TransitionState(name='Docked')
fusion = TransitionState(name='Fusion')
alpha = RateConstant(name="α", value=0.3, calcium_dependent=True)
beta = RateConstant(name="β", value=15)
transition1 = Transition(name='Transition 1',
rate_constant=alpha,
origin="Docked",
destination="Fusion")
transition2 = Transition(name='Transition 2',
rate_constant=beta,
origin="Fusion",
destination="Docked")
Add all objects to the model as follow:
model.add_transition_states([docked, fusion])
model.add_transitions([transition1, transition2])
Finally, initialize the model:
model.init()
Likewise, a stimulation protocol should be defined (if the experiment expects it) as follows:
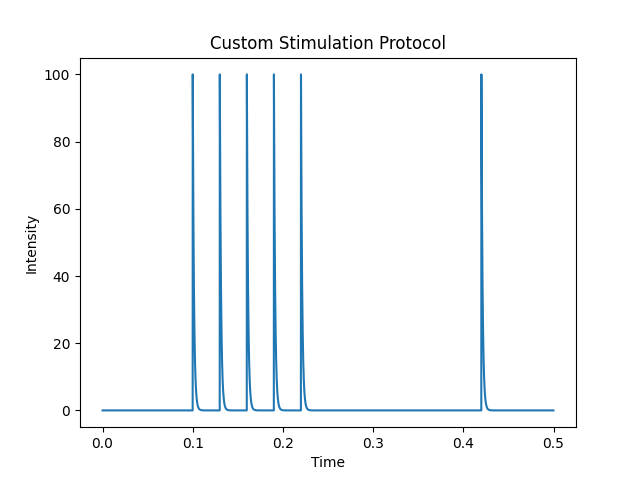
protocol = Stimulation(
conditional_stimuli=5,
period=0.03,
time_start_stimulation=0.1,
tau_stimulus=0.0013,
time_wait_test=0.2,
intensity_stimulus=100.0,
type_stimulus='exponential_decay',
name="Custom Stimulation Protocol")
The following lines show the time profile of the stimulation protocol:
import numpy as np
t = np.arange(0, 0.5, 0.0001)
protocol.plot(t)
Model Information (Optional)
The general model information can be obtained as follows:
model.get_info()
and run the file main.py:
$ python main.py
============== MODEL INFORMATION ===============
MODEL NAME: my-model
TOTAL VESICLES: 100
RESTING STATE: False
TRANSITION STATES VESICLES
- Docked: 100
- Fusion: 0
--------------------------------------------------
NAME TRANSITION: Transition 1
RATE CONSTANT NAME: α
RATE CONSTANT VALUE: 0.3 s⁻¹
CALCIUM-DEPENDENT: True
ORIGIN: Docked
DESTINATION: Fusion
--------------------------------------------------
NAME TRANSITION: Transition 2
RATE CONSTANT NAME: β
RATE CONSTANT VALUE: 15 s⁻¹
CALCIUM-DEPENDENT: False
ORIGIN: Fusion
DESTINATION: Docked
==================================================
The following lines allow you to visualize the graph of the model:
graph = model.get_graph()
graph.view()
Run It
The Solver object that simulates the time evolution of the model must be instantiated. Here, we use an implementation of the Gillespie Stochastic Algorithm (1977).
experiment = Solver(model=model, stimulation=protocol)
Before initiating the simulation, make sure to obtain the resting state of the model, from which all repetitions of the experiment are starting. This is achieved as follows:
experiment.resting_state()
With the following lines the experiment is ran. The results can be obtained and saved in a .csv file for further analysis.
experiment.run(repeat=1)
results = experiment.get_results(mean=True)
results.to_csv("results.csv", index=True)
Finally, the main.py file should be executed to perform the complete simulation:
$ python main.py
Changelog
- 0.1.3
- Fixed bug compatibility typing with python <= 3.8 versions.
- 0.1.2
- Added an option to include the individual events of the transitions in the results.
- A quantitative criterion was added to find the resting state automatically.
- Added the ability to include an external custom function when instantiating the Stimulation object.
- 0.1.1
- Added a progress bar when simulations are running.
- Fixed compatibility with pandas >=1.3.0 versions.
- Simplification of adding objects to the model. It is not necessary to
explicitly declare
KineticModel.add_rate_constants()method. - Added a AssertionError if the model is not initialized.
- Bug fixed in the number of vesicles when calling two or more times
KineticModel.init()method.
- 0.1.0
- Stable version released.
- 0.0.1
- Work in progress.
Contributing
-
If you are interested in contributing to the project, please follow these guidelines:
- Fork it (https://github.com/alexini-mv/kinetic-neurotransmission).
- Create your feature branch:
$ git checkout -b feature/fooBar
- Commit your changes:
$ git commit -am 'Add some fooBar'
- Push to the branch:
$ git push origin feature/fooBar
- Create a new Pull Request.
-
If you want to report a bug, please create a new issue here describing the error as clearly as possible.
Correspondence
Author: Alejandro Martínez-Valencia
Email: al.martinez.valencia@gmail.com
Citation
If you use our code for your research or scientific publication, we kindly ask you to refer to our work as follows:
- Martínez-Valencia A., Ramírez-Santiago G. and De-Miguel F.F. (2022) Dynamics of Neuromuscular Transmission Reproduced by Calcium-Dependent and Reversible Serial Transitions in the Vesicle Fusion Complex. Front. Synaptic Neurosci. 13:785361. DOI: 10.3389/fnsyn.2021.785361
License
Distributed under the GNU General Public License. See LICENSE for more information.
Interesting Links
- Perkel, Jeffrey M. (2022) How to fix your scientific coding errors. Nature 602, 172-173. DOI: 10.1038/d41586-022-00217-0.
- Pylint, a tool that checks for errors in Python code. Tutorials and documentation.
- Autopep8, a package automatically formats Python code to conform to the PEP 8 style guide. Documentation.
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.















