A humble collection of various everyday utility scripts from Team II IChO PAS.
Project description
zeetoo
A collection of various Python scripts created as a help in everyday work in Team II IChO PAS.
- Geting Started
- Running Scripts
- Description of modules
- analystex - converts plain text compound analyses to LaTeX
- backuper - simple automated backup tool for Windows
- confsearch - find conformers of given molecule using RDKit
- fixgvmol - correct .mol files created with GaussView software
- getcdx - extract all ChemDraw files embedded in .docx file
- gofproc - simple script for processing Gaussian output files
- sdf_to_gjf - save molecules from .sdf file as separate .gjf files
- Requirements
- License & Disclaimer
- Changelog
Getting Started
To use this collection of scripts you will need a Python 3 interpreter. You can download an installer of latest version from python.org (a shortcut to direct download for Windows: Python 3.7.4 Windows x86 executable installer).
The easiest way to get zeetoo up and running is to run pip install zeetoo in the command line*.
Alternatively, you can download this package as zip file using 'Clone or download' button on this site.
Unzip the package and from the resulting directory run python setup.py install
in the command line*.
And that's it, you're ready to go!
* On windows you can reach command line by right-clicking inside the directory while holding Shift and then choosing "Open PowerShell window here" or "Open command window here".
Running Scripts
Command Line Interface
All zeetoo functionality is available from command line.
After installation of the package each module can be accessed with use of
zeetoo [module_name] [parameters].
For more information run zeetoo --help to see available modules or
zeetoo [module_name] --help to see the help page for specific module.
Python API
Modules contained in zeetoo may also be used directly from python. This section will be supplemented with details on this topic soon.
Graphical User Interface
A simple graphical user interface (GUI) is available for backuper script. Please refer to the backuper section for details. GUIs for other modules will probably be available in near future.
Description of Modules
analystex
Parses plain text file containing list of analyses of chemical compounds and saves it in LaTeX format.
Resulting LaTeX code requires siunitx package and chemmacros package with 'spectroscopy' option.
Input file must contain following data entries in specified order, one entry (type of analysis) per line:
- compound's ID,
- compound's name,
- yield (Yield: XX%, compounds form),
- 1HNMR (1H NMR ( MHz, <solvent_as_formula>) δ = <shift/range> (, H), comma separated),
- 13CNMR (13C NMR ( MHz, <solvent_as_formula>) δ = , comma separated),
- IR peaks (IR () cm-1, comma separated),
- HRMS (HRMS () for : found – , calculated – ),
- specific rotation (α = (c = , solv. )),
- melting point (m.p. – ) (optional).
Data sets should be separated by an empty line.
backuper
A simple Python script for scheduling and running automated backup. Essentially, it copies specified files and directories to specified location with regard to date of last modification of both, source file and existing copy:
- if source file is newer than backup version, the second will be overridden;
- if both files have the same last modification time, file will not be copied;
- if backup version is newer, it will be renamed to "oldname_last-modification-time" and source file will be copied, preserving both versions.
After creating a specification for backup job (that is, specifying backup destination and files that should be copied; these information are stored in .ini file), it may be run manually or scheduled. Scheduling is currently available only on Windows, as it uses build-in Windows task scheduler. It is important to remember, that this is not a version control software. Only lastly copied version is stored. A minimal graphical user interface for this script is available (see below).
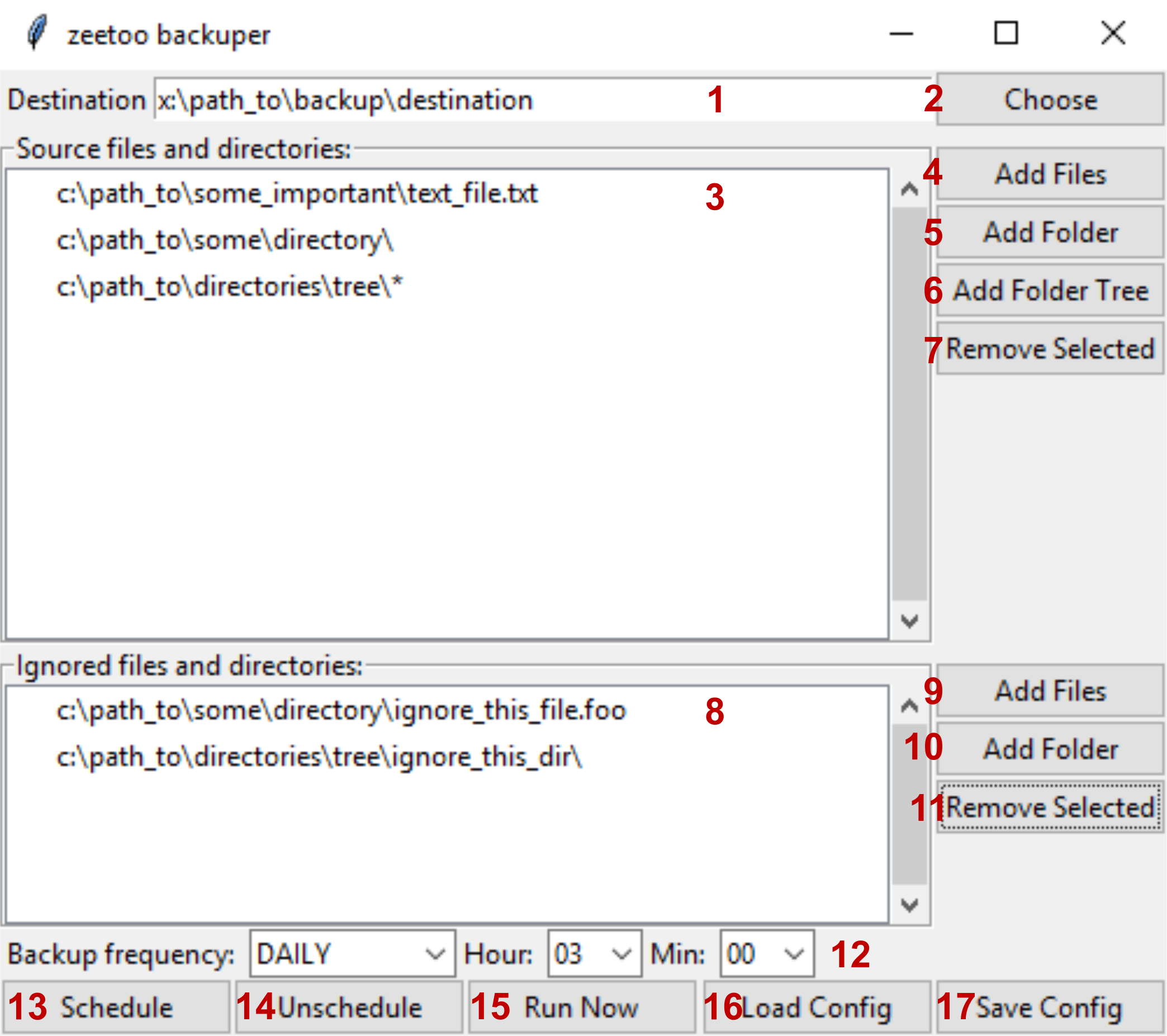
graphical user interface for backuper module
To start up the graphical user interface (GUI) run zeetoo backuper_gui in the command line.
If you've downloaded the whole package manually, you may also double-click on start_gui.bat file.
A window similar to the one below should appear.
Further you'll find description of each element of this window.
- This field shows path to backup's main directory. Files will be copied there. You can change this field directly or by clicking 2.
- Choose backup destination directory using graphical interface.
- All files and directories that are meant to be backuped are listed here. It will be called 'source' from now on. For more details read 4-7.
- Add file or files to source. Files will be shown in 3 as line without slash character at the end. Each file will be copied to the directory of the same name as directory it is located in; in example shown above it would be 'x:\path_to\backup\destination\some_important\text_file.text'.
- Add a directory to source. Directories will be shown in 3 as line with slash character at the end. All files (but not subdirectories!) present in this directory will be copied to directory with same name.
- Add a directory tree to source. Trees will be shown in 3 as line with slash and star characters at the end. The whole content of chosen directory will be copied, including all files and subdirectories.
- Remove selected path from source.
- All files and directories marked as ignored will be shown here. Ignored files and directories won't be copied during backup, even if they are inside source directory or tree, or listed as source.
- Add file or files to ignored.
- Add directory to ignored.
- Remove selected item from list of ignored files and directories.
- Set how often backup should be run (once a day, once a week or once a month) and at what time.
- Schedule backup task according to specified guidelines. WARNING: this will also automatically save configuration file.
- Remove backup task scheduled earlier.
- Run backup task now, according to specified guidelines. Saving configuration to file not needed.
- Load configuration from specified file.
- Save configuration.
Configuration is stored in [User]/AppData/Local/zeetoo/backuper/config.ini file.
After scheduling backup task this file should not be moved.
It can be modified though, backup task will be done with this modified guidelines from now on.
Scheduling new backup task, even using different configuration file, will override previous task,
unless task_name in this file is specifically changed.
confsearch
Performs a conformational search on set of given molecules. Takes a .mol file (or number of them) as an input and saves a list of generated conformers to specified .sdf file. Some restriction on this process may be given: a number of conformers to generate, a minimum RMSD value, a maximum energy difference, a maximum number of optimization cycles, and a set of constraints for force field optimization.
fixgvmol
.mol files created with GaussView (GV5 at least) lack some information, namely a mol version and END line. Without it some programs might not be able to read such files. This script adds these pieces of information to .mol files missing them.
getcdx
Extracts all embedded ChemDraw files from a .docx document and saves it in a separate directory (which might be specified by user), using in-text description of schemes/drawings as file names. It may be specified if description of the scheme/drawing is located above or underneath it (the former is default). Finally, It may be specified how long filename should be.
gofproc
Extracts information about molecule energy and imaginary frequencies from given set of Gaussian output files with freq job performed. Extracted data might be written to terminal (stdout) or to specified .xlsx file (must not be opened in other programs) at the end of the file or appended to a row, based on name of the file parsed. Calculations, that did not converged are reported separately.
sdf_to_gjf
Writes molecules contained in an .sdf file to a set of .gjf files in accordance with the guidelines given by user.
Requirements
- getcdx module requires olefile package
- gofproc module requires openpyxl package
- confsearch module requires RDKit software
Please note, that the RDKit will not be installed automatically with this package. The recommended way to get RDKit software is through use of Anaconda Python distribution. Please refer to RDKit documentation for more information.
License & Disclaimer
See the LICENSE.txt file for license rights and limitations (MIT).
Changelog
v.0.1.4
- added
analystexscript - fixed "run now" function in backuper's GUI
v.0.1.3
- fixed sdf_to_gjf ignoring parameters "charge" and "multiplicity"
- supplemented sdf_to_gjf default values and help message
- fixed typo in sdf_to_gjf CLI ("sufix" -> "suffix")
- enabled specifying coordinates' precision in sdf_to_gjf
- enhanced handling of link0 commands by sdf_to_gjf
- removed filtering of explicitly provided non-.mol files in fixgvmol
v.0.1.2
- getcdx now changes characters forbidden in file names to "-" instead of raising an exception
- start_gui.bat should now work regardless its location
v.0.1.1
- fixed import errors when run as module
v.0.1.0
- initial release
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.