CLI and utilities for Genetic analysis and database interface
Project description
geneutils
This tool was originally inspired by the work here for dealing with nucleotide sequences and more tools will be added as needed. This toolbox requires python 3 and can be installed.
Table of contents
Installation
This assumes that you have native python 3 & pip installed in your system, you can test this by going to the terminal (or windows command prompt) and trying
python and then pip list
If you get no errors and you have python 3 or higher you should be good to go. To install geneutils you can install using two methods
pip install geneutils
or you can also try
pip install geneutils --user
or you can also try
git clone https://github.com/samapriya/geneutils.git
cd geneutils
python setup.py install
Installation is an optional step; the application can be also run directly by executing pydrop.py script. The advantage of having it installed is being able to execute porg as any command line tool. I recommend installation within virtual environment. If you don't want to install, browse into the geneutils folder and try python geneutils.py -h to get to the same result.
geneutils cli tools
This is a command line tool and it is designed to simply call the tools you need.
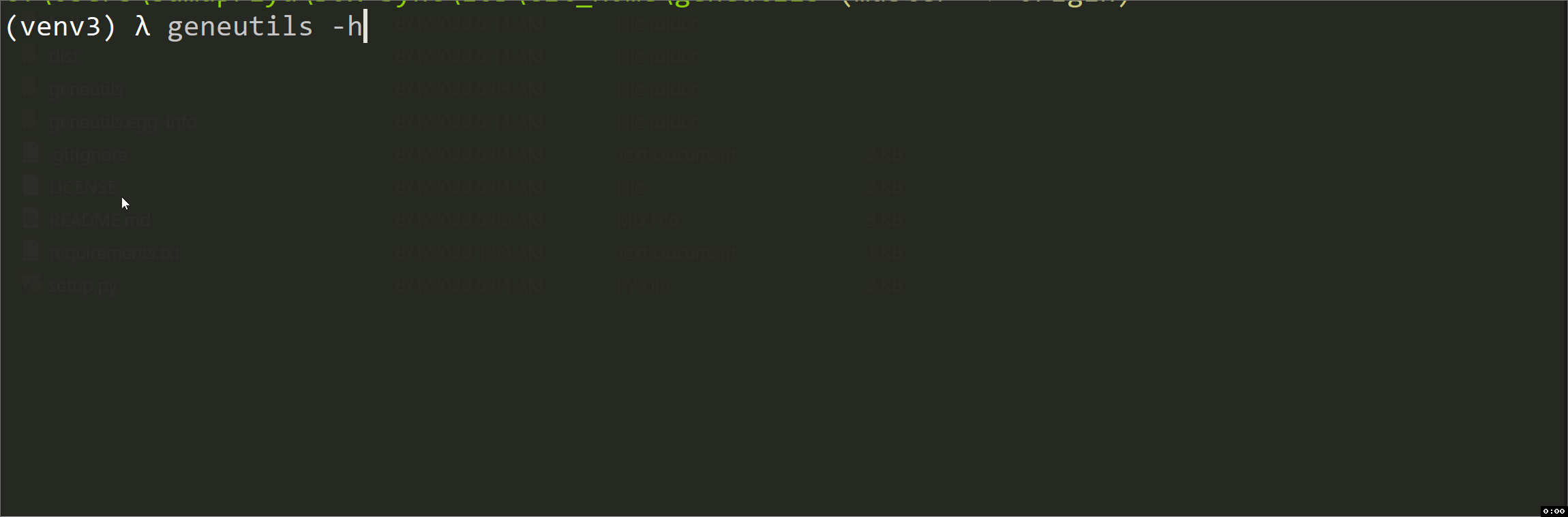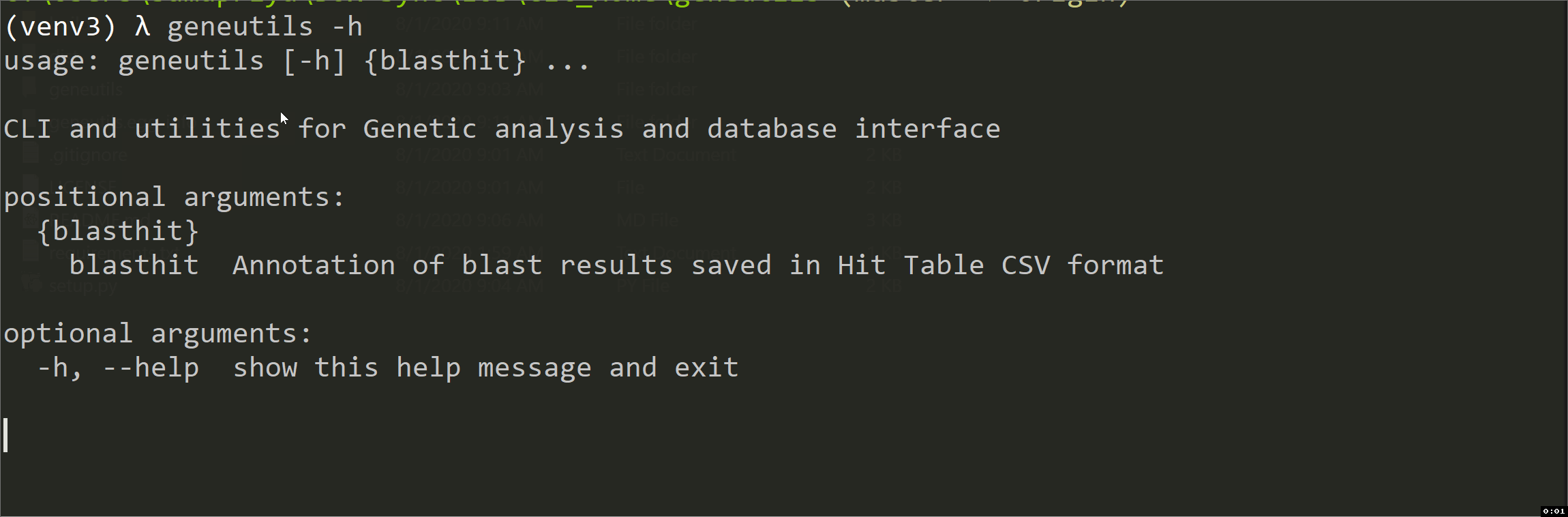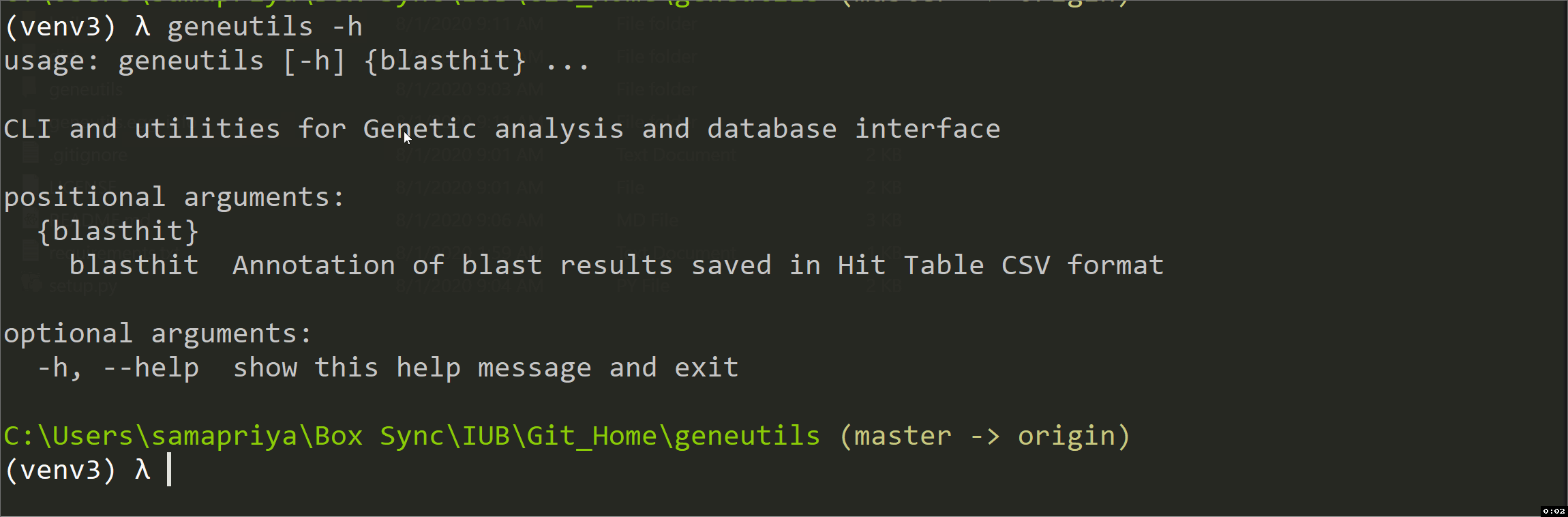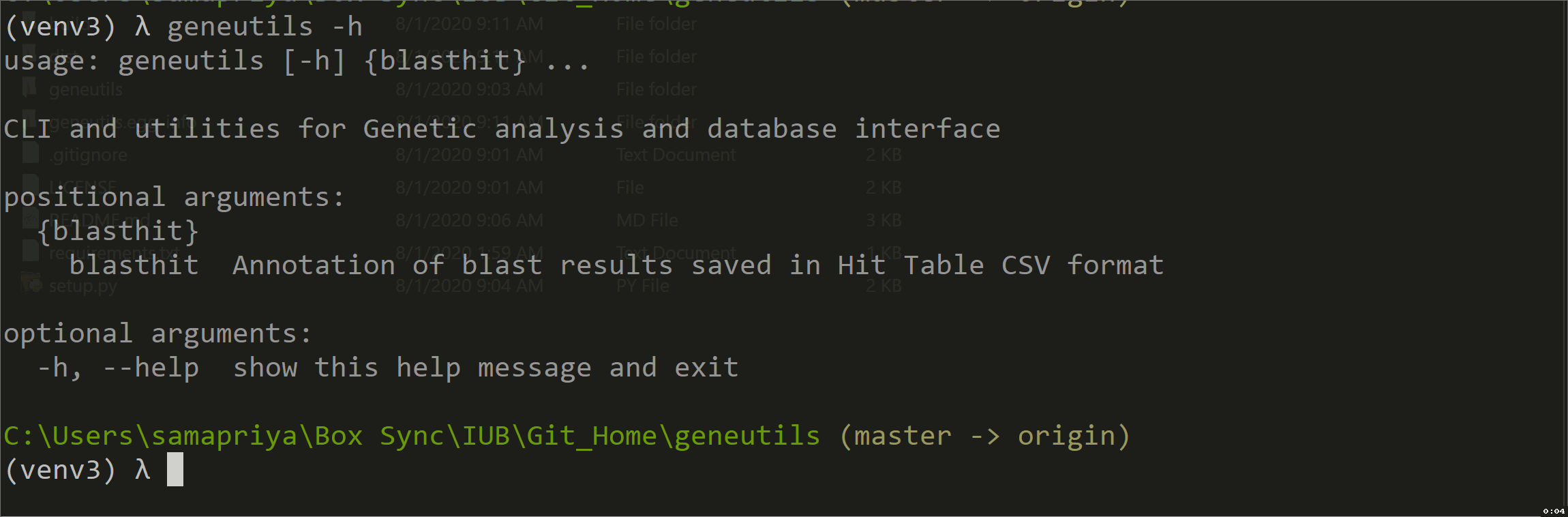
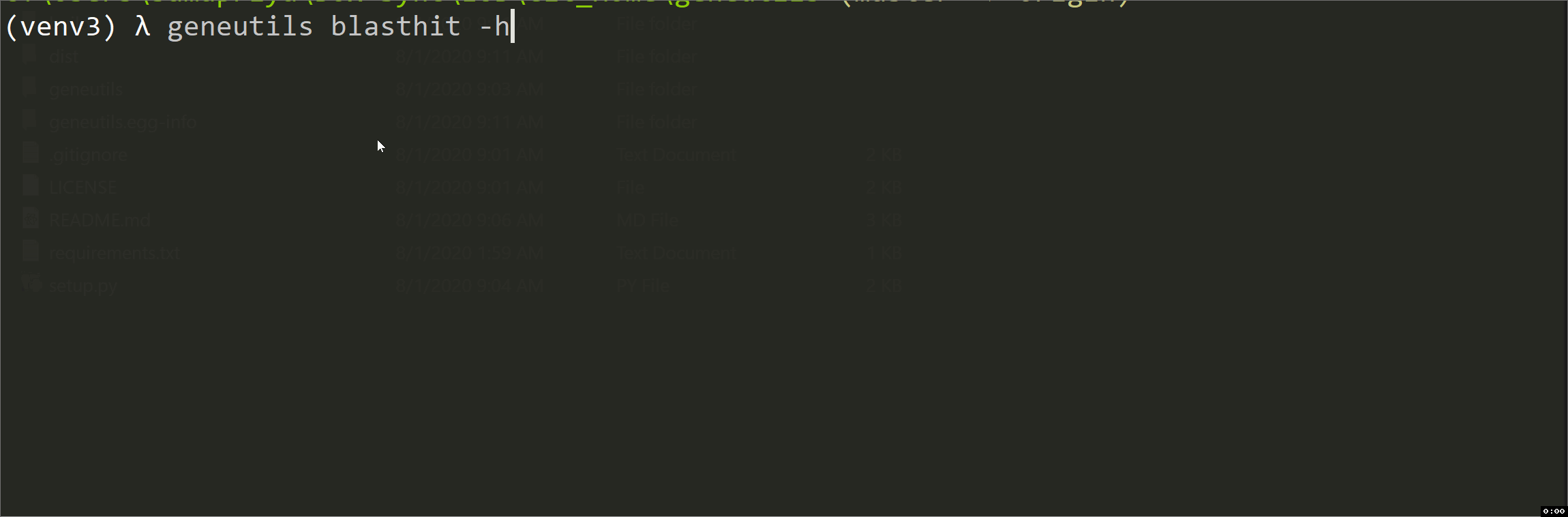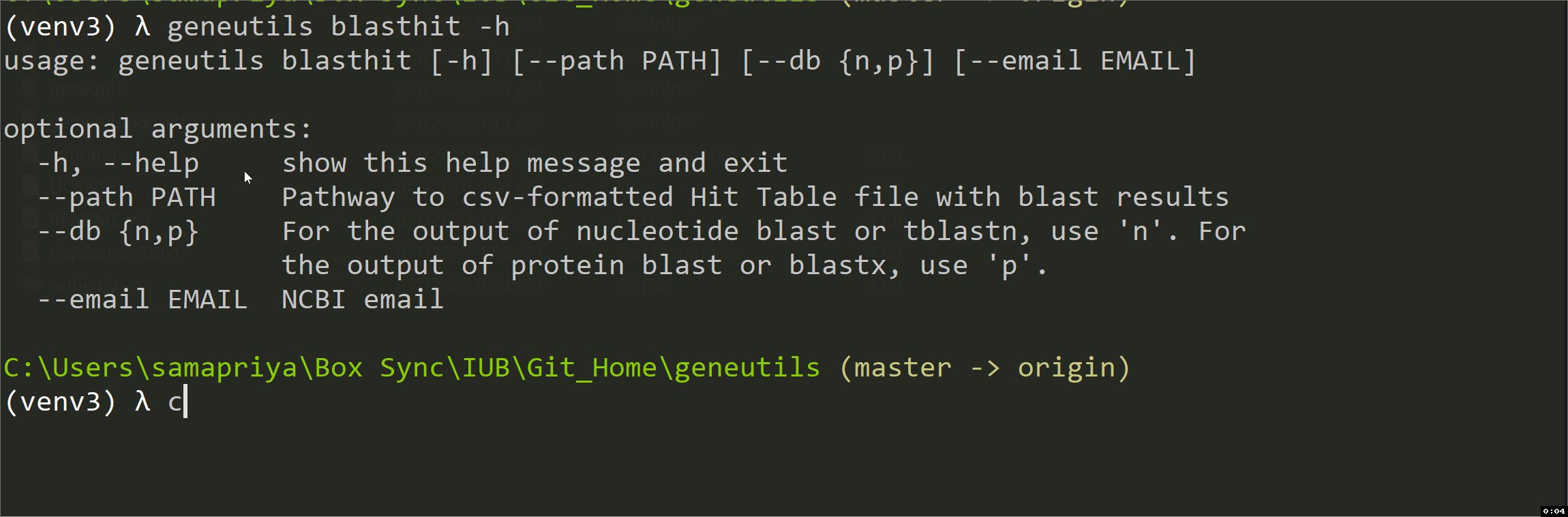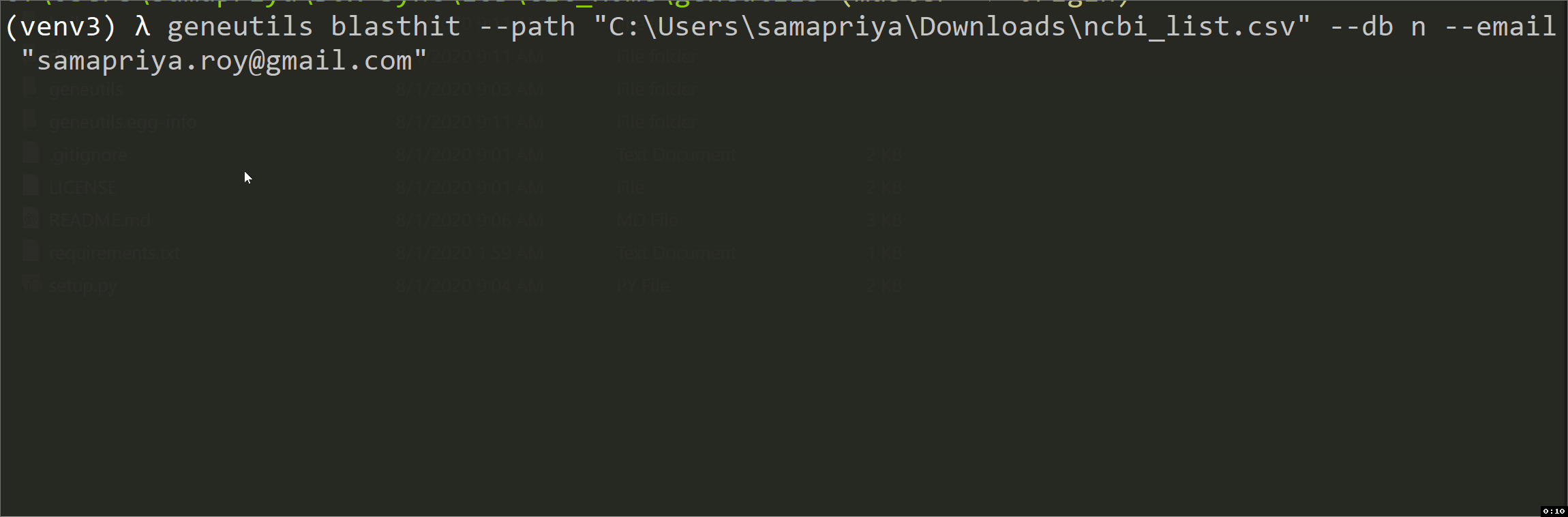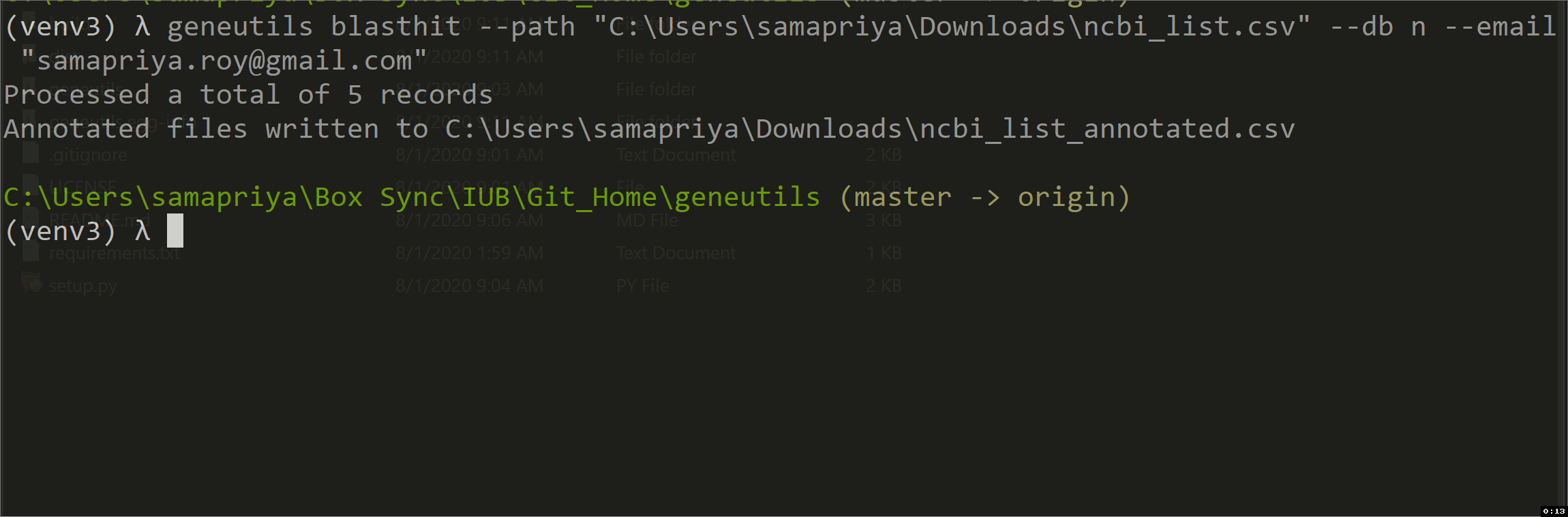
blasthit
This script is intended for taxonomic annotation of blast results (blastn, tblastn, blastp or blastx) saved in Hit Table CSV format where GenBank accession numbers are in the 4th column. It uses efetch function from Bio.Entrez package to get information about accessions from GenBank Nucleotide (Nuccore) or Protein databases. The output is an annotated CSV file "*_annotated.csv" with the following columns added:
- Record name
- Species name
- Full taxonomy
- Reference
- Date of update
| arguments | description |
|---|---|
| path | Pathway to csv-formatted Hit Table file with blastn results. Positional argument |
| db | For the output of nucleotide blast or tblastn, use n. For the output of protein blast or blastx, use p. Positional argument |
| Your NCBI email. Positional argument | |
| -h, --help | Show help message and exit. Optional argument |
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Hashes for geneutils-0.0.1-py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | ca45f87a40bcdd29adc63a976926d3418a18e0576873a4e342d540890044b1ce |
|
| MD5 | c4f90c2aadefc2c35762b14f39ac27b5 |
|
| BLAKE2b-256 | 56b5406e69b7ac621bf4c96e8c472405a09e2ccd2076de01d4d2f64416add94f |