Ancestry and haplotype aware simulation of genotypes and phenotypes for complex trait analysis
Project description
haptools
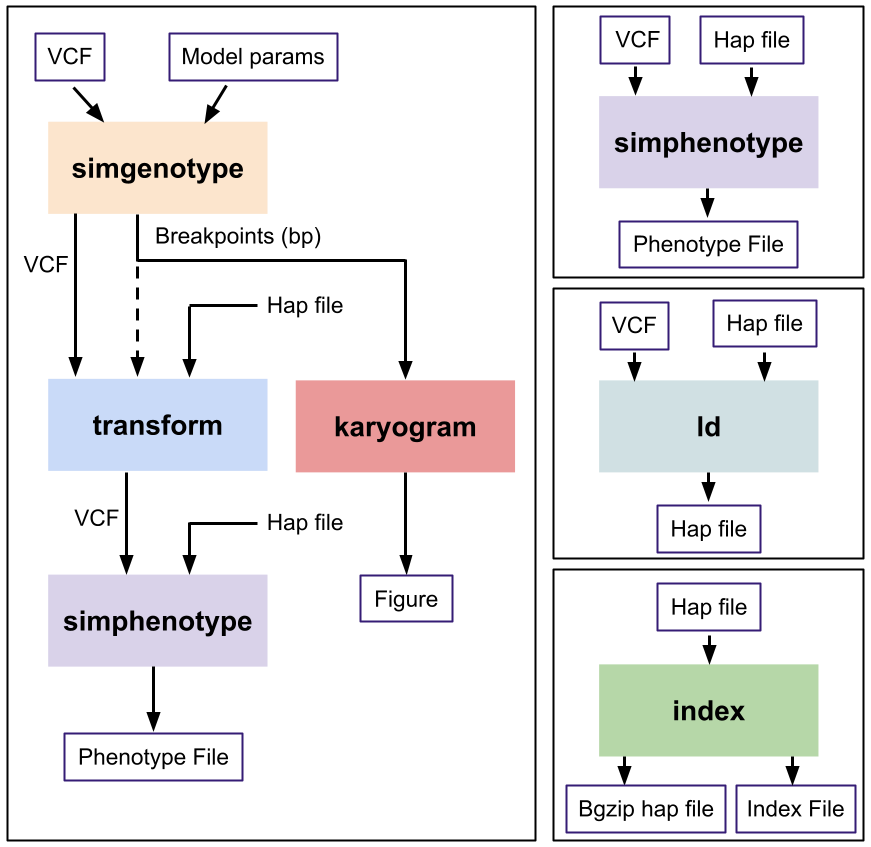
Haptools is a collection of tools for simulating and analyzing genotypes and phenotypes while taking into account haplotype information. Haptools supports fast simulation of admixed genomes (with simgenotype), visualization of admixture tracks (with karyogram), simulating haplotype- and local ancestry-specific phenotype effects (with transform and simphenotype), and computing a variety of common file operations and statistics in a haplotype-aware manner.
Homepage: https://haptools.readthedocs.io/
Visit our homepage for installation and usage instructions.
citation
There is an option to "Cite this repository" on the right sidebar of the repository homepage
Arya R Massarat, Michael Lamkin, Ciara Reeve, Amy L Williams, Matteo D’Antonio, Melissa Gymrek, Haptools: a toolkit for admixture and haplotype analysis, Bioinformatics, 2023;, btad104, https://doi.org/10.1093/bioinformatics/btad104
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.