Pipelines for detection epileptic spikes in MEG recording.
Project description
MEG-SPIKES
This repository contains functions for detecting, analyzing and evaluating epileptic spikes in MEG recording.
Installation
Optionally create a fresh virtual environment:
conda create -n megspikes pip python=3.7
The easiest way to install the package is using pip:
pip install megspikes
To install the latest version of the package, you should clone the repository and install all dependencies:
git clone https://github.com/MEG-SPIKES/megspikes.git
cd megspikes/
pip install .
Examples
Examples of how to use this package are prepared in the Jupyter Notebooks.
- 0_simulation.ipynb: simulation used to test this package and in other examples.
- 1_manual_pipeline.ipynb: localization of the irritative area for already detected (simulated) spikes.
- 2_aspire_alphacsc_pipepline.ipynb: full spikes detection pipeline and visualization of each step.
Documentation
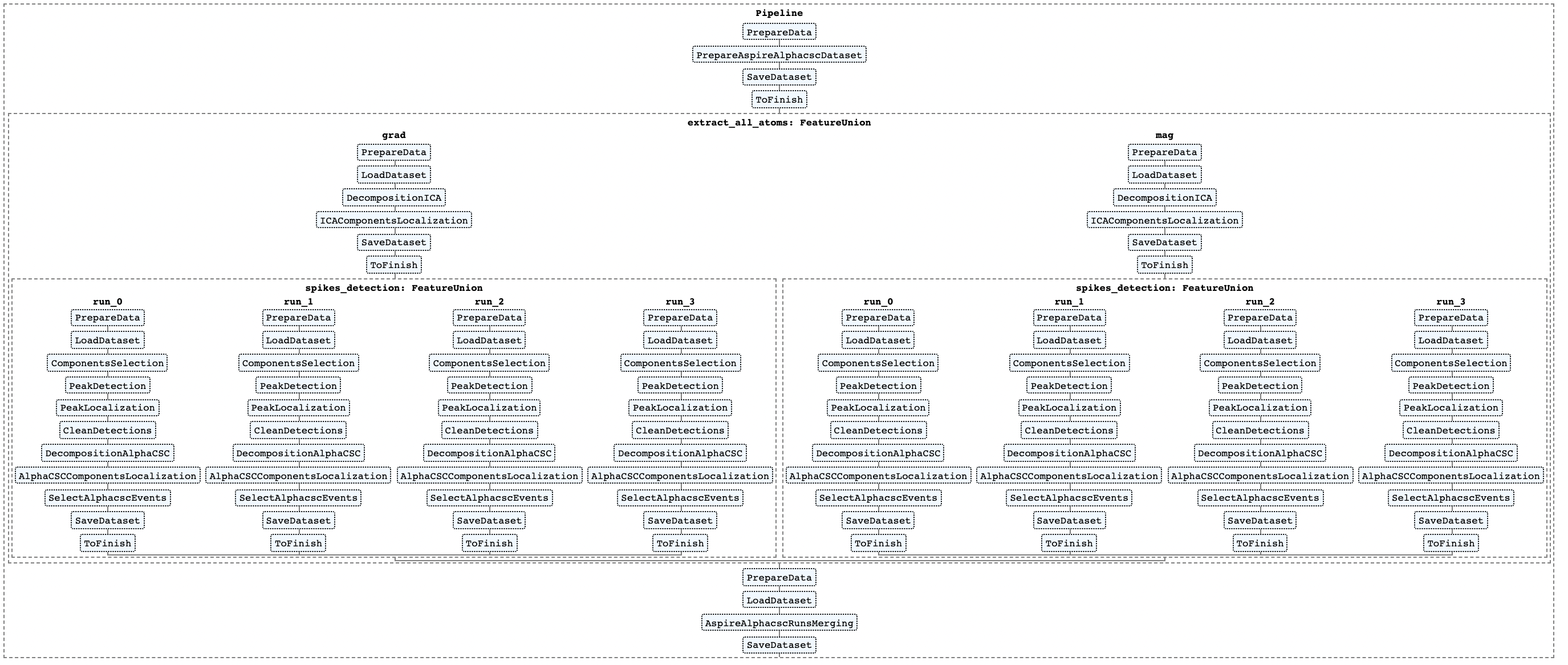
ASPIRE AlphaCSC pipeline
Full detection pipeline is presented on the figure below. The image was created using Scikit-learn Pipeline module.
To reproduce this picture see 2_aspire_alphacsc_pipepline.ipynb.
As is it depicted on the figure, ASPIRE-AlphaCSC pipeline includes the following main steps:
- ICA decomposition
- ICA components localization
- ICA components selection
- ICA peaks localization
- ICA peaks cleaning
- AlphaCSC decomposition
- AlphaCSC atoms localization
- AlphaCSC events selection
- AlphaCSC atoms merging
- AlphaCSC atoms goodness evaluation
- AlphaCSC atoms selection
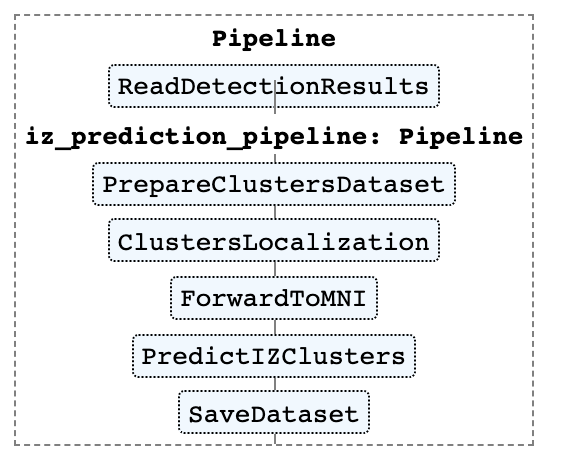
Clusters localization and the irritative area prediction
Irritative zone prediction pipeline is presented on the figure below. The image was created using Scikit-learn Pipeline module.
To reproduce this picture see 2_aspire_alphacsc_pipepline.ipynb and 1_manual_pipeline.ipynb.
Parameters
aspire_alphacsc_default_params.yml includes all default parameters that were used to run spike detection using combination of ASPIRE [2] and AlphaCSC [1].
clusters_default_params.yml describes all the parameters that were used for the irritative area prediction based on the detected events and their clustering.
Dependencies
Analysis
Data storing
Visualization
Testing
Contributing
All contributors are expected to follow the code of conduct.
References
[1] La Tour, T. D., Moreau, T., Jas, M., & Gramfort, A. (2018). Multivariate Convolutional Sparse Coding for Electromagnetic Brain Signals. ArXiv:1805.09654 [Cs, Eess, Stat]. http://arxiv.org/abs/1805.09654
[2] Ossadtchi, A., Baillet, S., Mosher, J. C., Thyerlei, D., Sutherling, W., & Leahy, R. M. (2004). Automated interictal spike detection and source localization in magnetoencephalography using independent components analysis and spatio-temporal clustering. Clinical Neurophysiology, 115(3), 508–522. https://doi.org/10.1016/j.clinph.2003.10.036
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.