Generate reports from metagenomics data
Project description
MicroView
MicroView, a reporting tool for taxonomic classification
MicroView agreggates results from taxonomic classification tools, such as Kaiju and Kraken, building an interactive HTML report with insightful visualizations.
Checkout the full documentation.
Quickstart
Install the package:
pip install microview
Alternatively, you can also install it with Conda:
conda install bioconda:microview
Go to your directory containing Kaiju/Kraken-style results and run:
microview -t .
Alternatively, if you have a CSV table defining result paths and contrasts, like this:
sample,group
result_1.tsv,group_one
result_2.tsv,group_two
...etc...
You can run MicroView like this:
microview -df contrast_table.csv -o report_with_table.html
Then, an HTML file named microview_report.html -
or the name you defined with the -o param -
should be available in your working directory,
try opening it with your browser!
Example report
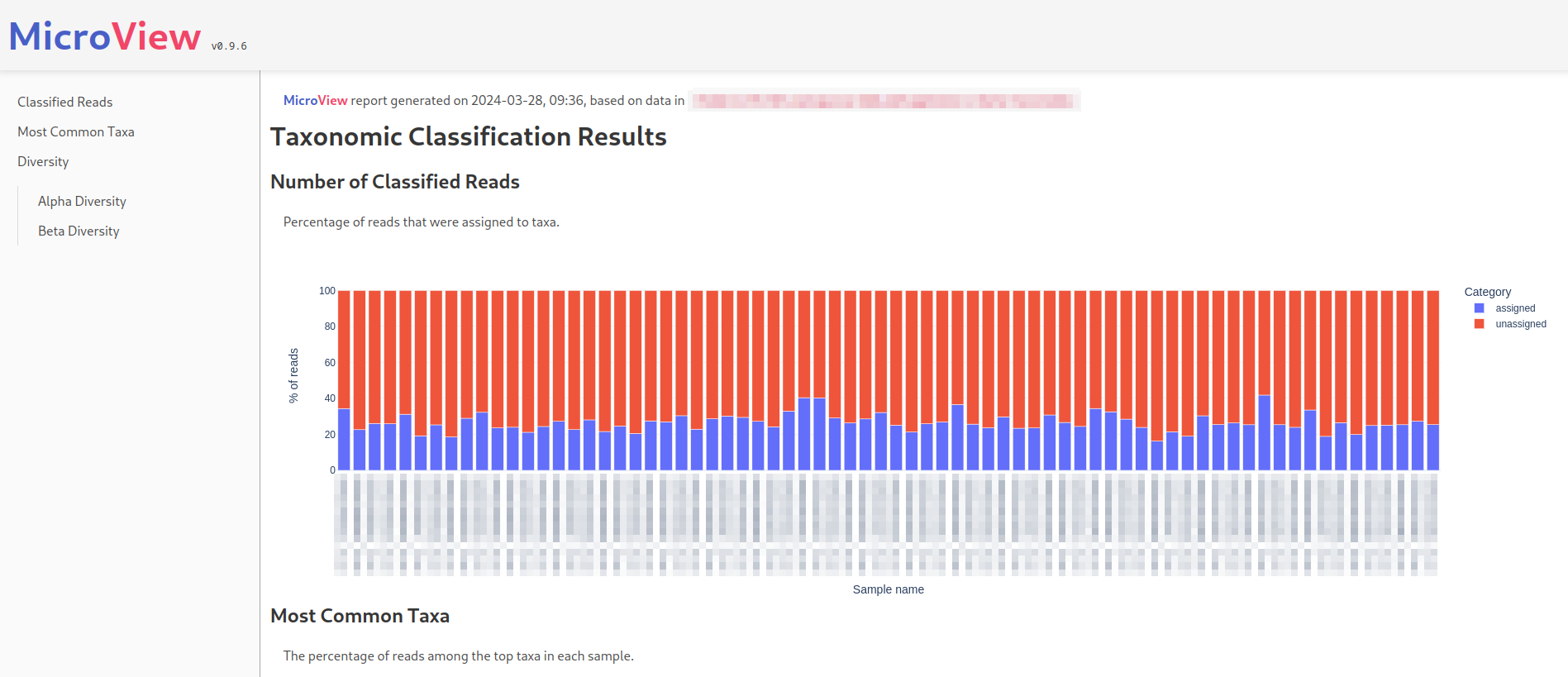
Here's what the beginning of a MicroView report looks like (sensitive information obscured):
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Hashes for MicroView-0.11.0-py2.py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | fa7b1d917552f0444627f41703114f341e4d8b65c1a12ca274316ebd75c24b3a |
|
| MD5 | 480c8e2040a7e10d78c47e034b6d186a |
|
| BLAKE2b-256 | 256759de8311b75aeade4c5934428d58aea4d749878e91b1266b9601f0510eb7 |