OntoGPT
Project description
OntoGPT
Introduction
OntoGPT is a Python package for the generation of Ontologies and Knowledge Bases using GPT. It is a knowledge extraction tool that uses a Large Language Models (LLMs) to extract semantic information from text.
This makes use of so-called instruction prompts in Large Language Models (LLMs) such as GPT-4.
Currently three different strategies for knowledge extraction have been implemented in the ontogpt package:
- SPIRES: Structured Prompt Interrogation and Recursive Extraction of Semantics
- Zero-shot learning (ZSL) approach to extracting nested semantic structures from text
- This approach takes two inputs - 1) LinkML schema 2) free text, and outputs knowledge in a structure conformant with the supplied schema in JSON, YAML, RDF or OWL formats
- Uses text-davinci-003 or gpt-3.5-turbo (gpt-4 untested)
- HALO: HAllucinating Latent Ontologies
- Few-shot learning approach to generating/hallucinating a domain ontology given a few examples
- Uses code-davinci-002
- SPINDOCTOR: Structured Prompt Interpolation of Narrative Descriptions Or Controlled Terms for Ontological Reporting
- Summarize gene set descriptions (pseudo gene-set enrichment)
- Uses text-davinci-003 or gpt-3.5-turbo (gpt-4 untested)
Pre-requisites
- Python 3.9+
- OpenAI account
- Optionally, BioPortal account (for grounding)
You will need to set both API keys using the Ontology Access Kit
poetry run runoak set-apikey -e openai <your openai api key>
poetry run runoak set-apikey -e bioportal <your bioportal api key>
Setup
For feature development and contributing to the package:
git clone https://github.com/monarch-initiative/ontogpt.git
cd ~/path/to/ontogpt
poetry install
To simply start using the package in your workspace:
pip install ontogpt
Examples
Strategy 1: Knowledge extraction using SPIRES
Input
Consider some text from one of the input files being used in the ontogpt test suite. You can find the text file here. You can download the raw file from the GitHub link to that input text file, or copy its contents over into another file, say, abstract.txt. An excerpt
The cGAS/STING-mediated DNA-sensing signaling pathway is crucial for interferon (IFN) production and host antiviral responses
... [snip] ...
The underlying mechanism was the interaction of US3 with β-catenin and its hyperphosphorylation of β-catenin at Thr556 to block its nuclear translocation ... ...
We can extract knowledge from the above text this into the GO pathway datamodel by running the following command:
Command
ontogpt extract -t gocam.GoCamAnnotations -i ~/path/to/abstract.txt
Note: The value accepted by the -t / --template argument is the base name of one of the LinkML schema / data model which can be found in the templates folder.
Output
The output returned from the above command is knowledge can be optionally redirected into an output file using the -o / --output.
The following is a small part of what the larger schema-compliant output looks like:
genes:
- HGNC:2514
- HGNC:21367
- HGNC:27962
- US3
- FPLX:Interferon
- ISG
gene_gene_interactions:
- gene1: US3
gene2: HGNC:2514
gene_localizations:
- gene: HGNC:2514
location: Nuclear
gene_functions:
- gene: HGNC:2514
molecular_activity: Transcription
- gene: HGNC:21367
molecular_activity: Production
...
Working Mechanism
- You provide an arbitrary data model, describing the structure you want to extract text into
- This can be nested (but see limitations below)
- Provide your preferred annotations for grounding
NamedEntityfields - OntoGPT will:
- Generate a prompt
- Feed the prompt to a language model (currently OpenAI GPT models)
- Parse the results into a dictionary structure
- Ground the results using a preferred annotator
Strategy 2: HALO
Documentation to come
Strategy 3: Gene Enrichment using SPINDOCTOR
Given a set of genes, OntoGPT can find similarities among them.
Ex.:
ontogpt enrichment -U tests/input/genesets/sensory-ataxia.yaml
The default is to use ontological gene function synopses (via the Alliance API).
- To use narrative/RefSeq summaries, use the
--no-ontological-synopsesflag - To run without any gene descriptions, use the
--no-annotationsflag
Features
Define your own extraction model using LinkML
There are a number of pre-defined LinkML data models already developed here - src/ontogpt/templates/ which you can use as reference when creating your own data models.
Define a schema (using a subset of LinkML) that describes the structure in which you want to extract knowledge from your text.
example custom linkml data model
```yaml classes: MendelianDisease: attributes: name: description: the name of the disease examples: - value: peroxisome biogenesis disorder identifier: true ## needed for inlining description: description: a description of the disease examples: - value: >- Peroxisome biogenesis disorders, Zellweger syndrome spectrum (PBD-ZSS) is a group of autosomal recessive disorders affecting the formation of functional peroxisomes, characterized by sensorineural hearing loss, pigmentary retinal degeneration, multiple organ dysfunction and psychomotor impairment synonyms: multivalued: true examples: - value: Zellweger syndrome spectrum - value: PBD-ZSS subclass_of: multivalued: true range: MendelianDisease examples: - value: lysosomal disease - value: autosomal recessive disorder symptoms: range: Symptom multivalued: true examples: - value: sensorineural hearing loss - value: pigmentary retinal degeneration inheritance: range: Inheritance examples: - value: autosomal recessive genes: range: Gene multivalued: true examples: - value: PEX1 - value: PEX2 - value: PEX3Gene:
is_a: NamedThing
id_prefixes:
- HGNC
annotations:
annotators: gilda:, bioportal:hgnc-nr
Symptom:
is_a: NamedThing
id_prefixes:
- HP
annotations:
annotators: sqlite:obo:hp
Inheritance:
is_a: NamedThing
annotations:
annotators: sqlite:obo:hp
```
- Prompt hints can be specified using the
promptannotation (otherwise description is used) - Multivalued fields are supported
- The default range is string — these are not grounded. Ex.: disease name, synonyms
- Define a class for each
NamedEntity - For any
NamedEntity, you can specify a preferred annotator using theannotatorsannotation
We recommend following an established schema like BioLink Model, but you can define your own.
Next step is to compile the schema. For that, you should place the schema YAML in the directory src/ontogpt/templates/. Then, run the make command at the top level. This will compile the schema to Python (Pydantic classes).
Once you have defined your own schema / data model and placed in the correct directory, you can run the extract command.
Ex.:
ontogpt extract -t mendelian_disease.MendelianDisease -i marfan-wikipedia.txt
Multiple levels of nesting
Currently no more than two levels of nesting are recommended.
If a field has a range which is itself a class and not a primitive, it will attempt to nest.
Ex. the gocam schema has an attribute:
attributes:
...
gene_functions:
description: semicolon-separated list of gene to molecular activity relationships
multivalued: true
range: GeneMolecularActivityRelationship
The range GeneMolecularActivityRelationship has been specified inline, so it will nest.
The generated prompt is:
gene_functions : <semicolon-separated list of gene to molecular activities relationships>
The output of this is then passed through further SPIRES iterations.
Text length limit
Currently SPIRES must use text-davinci-003, which has a total 4k token limit (prompt + completion).
You can pass in a parameter to split the text into chunks. Returned results will be recombined automatically, but more experiments need to be done to determined how reliable this is.
Schema tips
It helps to have an understanding of the LinkML schema language, but it should be possible to define your own schemas using the examples in src/ontogpt/templates as a guide.
OntoGPT-specific extensions are specified as annotations.
You can specify a set of annotators for a field using the annotators annotation.
Ex.:
Gene:
is_a: NamedThing
id_prefixes:
- HGNC
annotations:
annotators: gilda:, bioportal:hgnc-nr, obo:pr
The annotators are applied in order.
Additionally, when performing grounding, the following measures can be taken to improve accuracy:
- Specify the valid set of ID prefixes using
id_prefixes - Some vocabularies have structural IDs that are amenable to regexes, you can specify these using
pattern - You can make use of
values_fromslot to specify a Dynamic Value Set- For example, you can constrain the set of valid locations for a gene product to be subclasses of
cellular_componentin GO orcellin CL
- For example, you can constrain the set of valid locations for a gene product to be subclasses of
Ex.:
classes:
...
GeneLocation:
is_a: NamedEntity
id_prefixes:
- GO
- CL
annotations:
annotators: "sqlite:obo:go, sqlite:obo:cl"
slot_usage:
id:
values_from:
- GOCellComponentType
- CellType
enums:
GOCellComponentType:
reachable_from:
source_ontology: obo:go
source_nodes:
- GO:0005575 ## cellular_component
CellType:
reachable_from:
source_ontology: obo:cl
source_nodes:
- CL:0000000 ## cell
OWL Exports
The extract command will let you export the results as OWL axioms, utilizing linkml-owl mappings in the schema.
Ex.:
ontogpt extract -t recipe -i recipe-spaghetti.txt -o recipe-spaghetti.owl -O owl
src/ontogpt/templates/recipe.yaml is an example schema that uses linkml-owl mappings.
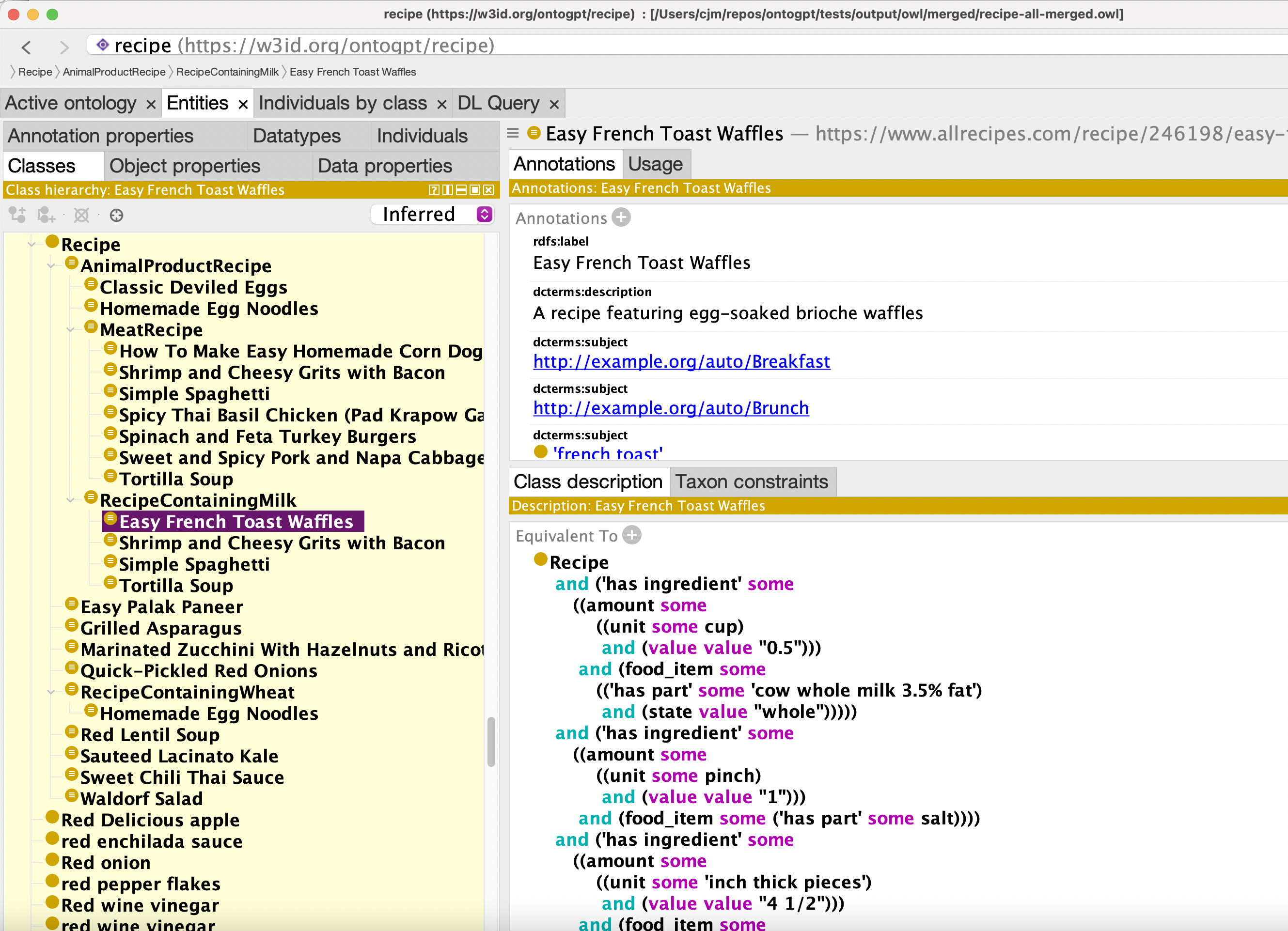
See the Makefile for a full pipeline that involves using robot to extract a subset of FOODON and merge in the extracted results. This uses recipe-scrapers.
OWL output: recipe-all-merged.owl
Classification:
Web Application Setup
There is a bare bones web application for running OntoGPT and viewing results.
Install the required dependencies by running the following command:
poetry install -E web
Then run this command to start the web application:
poetry run web-ontogpt
Note: The agent running uvicorn must have the API key set, so for obvious reasons don't host this publicly without authentication, unless you want your credits drained.
OntoGPT Limitations
- Non-deterministic
- This relies on an existing LLM, and LLMs can be fickle in their responses
- Coupled to OpenAI
- You will need an OpenAI account to use their API. In theory any LLM can be used but in practice the parser is tuned for OpenAI's models
SPINDOCTOR web app
To start:
poetry run streamlit run src/ontogpt/streamlit/spindoctor.py
Citation
SPIRES is described further in: Caufield JH, Hegde H, Emonet V, Harris NL, Joachimiak MP, Matentzoglu N, et al. Structured prompt interrogation and recursive extraction of semantics (SPIRES): A method for populating knowledge bases using zero-shot learning.
arXiv publication: http://arxiv.org/abs/2304.02711
Contributing
Contributions on recipes to test welcome from anyone! Just make a PR here. See this list for accepted URLs
Acknowledgements
We gratefully acknowledge Bosch Research for their support of this research project.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.














