Minimum-Distortion Embedding
Project description
PyMDE

PyMDE is a Python library for computing vector embeddings for finite sets of items, such as images, biological cells, nodes in a network, or any other abstract object.
What sets PyMDE apart from other embedding libraries is that it provides a simple but general framework for embedding, called Minimum-Distortion Embedding (MDE). With MDE, it is easy to recreate well-known embeddings and to create new ones, tailored to your particular application. PyMDE is competitive in runtime with more specialized embedding methods. With a GPU, it can be even faster.
PyMDE can be enjoyed by beginners and experts alike. It can be used to:
- reduce the dimensionality of vector data;
- generate feature vectors;
- visualize datasets, small or large;
- draw graphs in an aesthetically pleasing way;
- find outliers in your original data;
- and more.
PyMDE is very young software, under active development. If you run into issues, or have any feedback, please reach out by filing a Github issue.
This README is a crash course on how to get started with PyMDE. The full documentation is still under construction.
Installation
PyMDE is available on the Python Package Index. Install it with
pip install pymde
Getting started
Getting started with PyMDE is easy. For embeddings that work out-of-the box, we provide two main functions:
pymde.preserve_neighbors
which preserves the local structure of original data, and
pymde.preserve_distances
which preserves pairwise distances or dissimilarity scores in the original data.
Arguments. The input to these functions is the original data, represented
either as a data matrix in which each row is a feature vector, or as a
(possibly sparse) graph encoding pairwise distances. The embedding dimension is
specified by the embedding_dim keyword argument, which is 2 by default.
Return value. The return value is an MDE object. Calling the embed()
method on this object returns an embedding, which is a matrix
(torch.Tensor) in which each row is an embedding vector. For example, if the
original input is a data matrix of shape (n_items, n_features), then the
embedding matrix has shape (n_items, embeddimg_dim).
We give examples of using these functions below.
Preserving neighbors
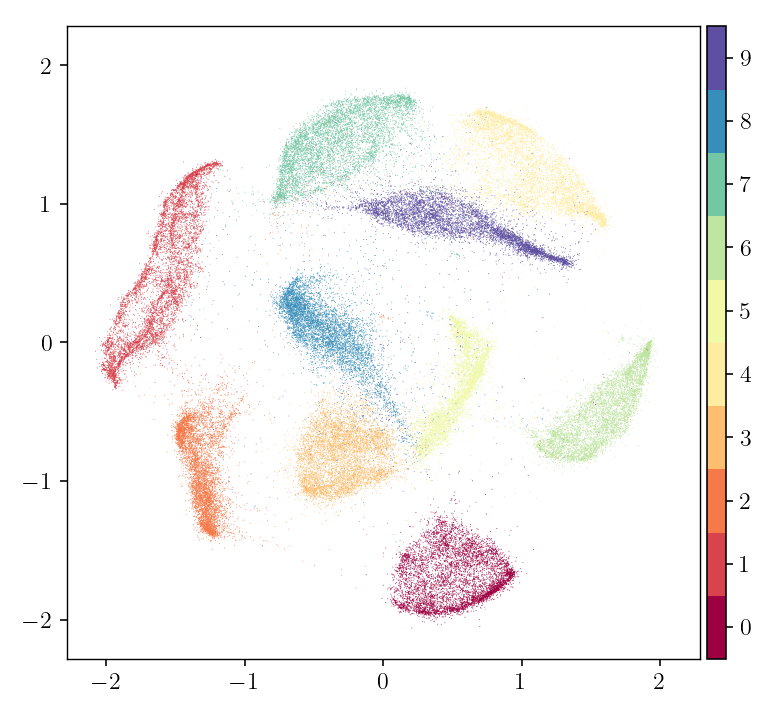
The following code produces an embedding of the MNIST dataset (images of
handwritten digits), in a fashion similar to LargeVis, t-SNE, UMAP, and other
neighborhood-based embeddings. The original data is a matrix of shape (70000, 784), with each row representing an image.
import pymde
mnist = pymde.datasets.MNIST()
embedding = pymde.preserve_neighbors(mnist.data).embed()
pymde.plot(embedding, color_by=mnist.attributes['digits'])
Unlike most other embedding methods, PyMDE can compute embeddings that satisfy constraints. For example:
embedding = pymde.preserve_neighbors(mnist.data, constraint=pymde.Standardized()).embed()
pymde.plot(embedding, color_by=mnist.attributes['digits'])
The standardization constraint enforces the embedding vectors to be centered and have uncorrelated features.
Preserving distances
The function pymde.preserve_distances is useful when you're more interested
in preserving the gross global structure instead of local structure.
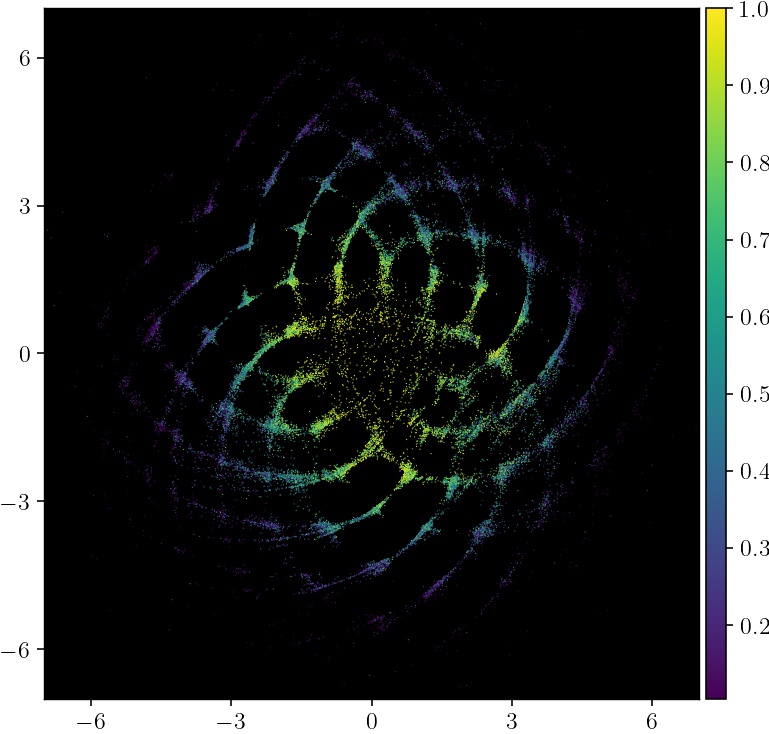
Here's an example that produces an embedding of an academic coauthorship network, from Google Scholar. The original data is a sparse graph on roughly 40,000 authors, with an edge between authors who have collaborated on at least one paper.
import pymde
google_scholar = pymde.datasets.google_scholar()
embedding = pymde.preserve_distances(google_scholar.data).embed()
pymde.plot(embedding, color_by=google_scholar.attributes['coauthors'], color_map='viridis', background_color='black')
More collaborative authors are colored brighter, and are near the center of the embedding.
MDE
The functions pymde.preserve_neighbors and pymde.preserve_distances from
the previous section both returned instances of the MDE class. For simple
use cases, you can just call the embed() method on the returned instance and
use the resulting embedding.
For more advanced use cases, such as creating custom embeddings, debugging
embeddings, and looking for outliers, you'll need to understand what an MDE
object represents.
We'll go over the basics of the MDE class in this section.
Hello, World
An MDE instance represents an "MDE problem", whose solution is an embedding. An MDE problem has 5 parts:
- the number of items being embedded
- the embedding dimension
- a list of edges
- distortion functions
- a constraint
We'll explain what these parts are using a simple example: embedding the nodes of a cycle graph into 2 dimensions.
Number of items. We'll embed a cycle graph on 5 nodes. We have 5 items (the
nodes we are embedding), labeled 0, 1, 2, 3, and 4.
Embedding dimension. The embedding dimension is 2.
Edges. Every MDE problem has a list of edges (i.e., pairs) (i, j), 0 <= i < j < n_items. If an edge (i, j) is in this list, it means we have an
opinion on the relationship between items i and j (e.g., we know that they
are similar).
In our example, we are embedding a cycle graph, and we know that adjacent nodes are similar. We choose the edges as
edges = torch.tensor([[0, 1], [1, 2], [2, 3], [3, 4], [0, 4]])
Notice that edges is a torch.Tensor of shape (n_edges, 2).
Distortion functions. The relationships between items are articulated by
distortion functions (or loss functions). Each distortion function is
associated with one pair (i, j), and it maps the Euclidean distance d_{ij}
between the embedding vectors for items i and j to a scalar distortion
(or loss). The goal of an MDE problem is to find an embedding that minimizes
the average distortion, across all edges.
In this example, for each edge (i, j), we'll use the distortion function
w_{ij} d_{ij}**3, where w_{ij} is a scalar weight. We'll set all the
weights to one for simplicity. This means we'll penalize large distances, and
treat all edges equally.
distortion_function = pymde.penalties.Cubic(weights=torch.ones(edges.shape[0]))
In general, the constructor for a penalty takes a torch.Tensor of weights,
with weights[k] giving the weight associated with edges[k].
Constraint. In our simple example, the embedding will collapse to 0 if it is unconstrained (since all the weights are positive). We need a constraint that forces the embedding vectors to spread out.
The constraint we will impose is called a standardization constraint. This constraint requires the embedding vectors to be centered, and to have uncorrelated feature columns. This forces the embedding to spread out.
We create the constraint with:
constraint = pymde.Standardized()
Creating the MDE problem. Having specified the number of items, embedding dimension, edges, distortion function, and constraint, we can now create the embedding problem:
import pymde
mde = pymde.MDE(
n_items=5,
embedding_dim=2,
edges=torch.tensor([[0, 1], [1, 2], [2, 3], [3, 4], [0, 4]]),
distortion_function=pymde.penalties.Cubic(weights=torch.ones(edges.shape[0])),
constraint=pymde.Standardized())
Computing an embedding. Finally, we can compute the embedding by calling
the embed method:
embedding = mde.embed()
print(embedding)
prints
tensor([[ 0.5908, -1.2849],
[-1.0395, -0.9589],
[-1.2332, 0.6923],
[ 0.2773, 1.3868],
[ 1.4046, 0.1648]])
(This method returns the embedding, and it also saves it in the X attribute
of the MDE instance (mde.X).)
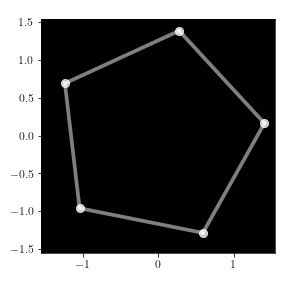
Since this embedding is two-dimensional, we can also plot it:
mde.plot(edges=mde.edges)
The embedding vectors are the dots in this image. Notice that we passed the
list of edges to the plot method, to visualize each edge along with the
vectors.
Summary. This was a toy example, but it showed all the main parts of an MDE problem:
n_items: the number of things ("items") being embeddedembedding_dim: the dimension of the embedding (e.g., 1, 2, or 3 for visualization, or larger for feature generation)edges: atorch.Tensor, of shape(n_edges, 2), in which each row is a pair(i, j).distortion_function: a vectorized distortion function, mapping atorch.Tensorcontaining then_edgesembedding distances to atorch.Tensorof distortions of the same length.constraint: a constraint instance, such aspymde.Standardized(), orNonefor unconstrained embeddings.
Next, we'll learn more about distortion functions, including how to write custom distortion functions.
Distortion functions
The vectorized distortion function is just any callable that maps a
torch.Tensor of embedding distances to a torch.Tensor of distortions, using
PyTorch operations. The callable must have the signature
distances: torch.Tensor[n_edges] -> distortions: torch.Tensor[n_edges]
Both the input and the return value must have shape (n_edges,), where
n_edges is the number of edges in the MDE problem. The entry distances[k]
is the embedding distance corresponding to the kth edge, and the entry
distortions[k] is the distortion for the kth edge.
As a simple example, the cubic penalty we used in the Hello, World example can be implemented as
def distortion_function(distances):
return distances ** 3
Distortion functions usually come as one of two types: they are either derived from scalar weights (one weight per edge) or deviations (also one per edge). PyMDE provides a library of distortion functions derived from weights, as well as a library of distortion functions derived from original deviations. These functions are called penalties and losses respectively.
Penalties. Distortion functions derived from weights have the general form
distortion[k] := w[k] penalty(distances[k]), where w[k] is the weight
associated with the k-th edge and penalty is a penalty function. Penalty
functions are increasing functions. A positive weight
indicates that the items in the kth edge are similar, and a negative weight
indicates that they are dissimilar. The larger the magnitude of the weight, the
more similar or dissimilar the items are.
A number of penalty functions are available in the pymde.penalties module.
Losses. Distortion functions derived from original deviations have the
general form distortion[k] = loss(deviations[k], distances[k]), where
deviations[k] is a positive number conveying how dissimilar the items in
edges[k] are; the larger deviations[k] is, the more dissimilar the items
are. The loss function is 0 when deviations[k] == distances[k], and
nonnegative otherwise.
A number of loss functions are available in the pymde.losses module.
Preprocessing data
The preserve_neighbors and preserve_distances functions create MDE problems
based on weights and original deviations respectively, preprocessing the
original data to obtain the weights or distances.
A number of preprocessors are provided in the pymde.preprocess module.
Example notebooks
The best way to learn more about PyMDE is to try it out, using the example notebooks as a starting point.
Citing
To cite our work, please use the following BibTex entry.
@article{agrawal2020minimum,
author = {Agrawal, Akshay and Ali, Alnur and Boyd, Stephen},
title = {Minimum-Distortion Embedding},
year = {2020},
}
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distributions
Hashes for pymde-0.1.0-cp38-cp38-win_amd64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | a47c9f44537d25cdb66a35f8e0937886d2f13503c39aba8387dd249d3fa13934 |
|
| MD5 | 612db60f9d14b5f0baef53b0c4ce0cb0 |
|
| BLAKE2b-256 | c9dd3c52b228cdbba04ca649f8add20cfab10e6cfce06acb645ad3d98af626f3 |
Hashes for pymde-0.1.0-cp38-cp38-manylinux2010_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 85f65a36b29bbe97b9b1ae97d7deaed4314ed1f4b0e4107bf8569cdc10d8c8db |
|
| MD5 | 941616d242da25c015b82fae78d33aa5 |
|
| BLAKE2b-256 | f0cdda64fb556d0a7ed361847b2a88e1af3c22cca74581e049d72d0d0928e509 |
Hashes for pymde-0.1.0-cp38-cp38-manylinux2010_i686.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | c3b953186737e21e72f063d7d5482a0f189feb7b3da81bc913ff04bf4da7b13f |
|
| MD5 | 519aa0985bbf907eff95e897c202f9b3 |
|
| BLAKE2b-256 | 5c93312c77cde586a477bb19c6f4c614d7d14a12acfa6333e3440b9687fad0ba |
Hashes for pymde-0.1.0-cp38-cp38-manylinux1_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | b82c6f4399b64f3fdc0bf74b9cf855c9f456979558a3d7e7f2696384adfa25f8 |
|
| MD5 | a47198bd1bf0d341cf2e1cd0bbe0d609 |
|
| BLAKE2b-256 | 8c5b0ea635589edc583eb18313307b2d9800fa4f67a7d99f12e998ec31c6b7b3 |
Hashes for pymde-0.1.0-cp38-cp38-manylinux1_i686.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 698ae0c055fe92dca13b26f6a346e40beaa5e26aaaafbb911b87058951f436a8 |
|
| MD5 | ba9475d334a6d136420665a2e3dc9db2 |
|
| BLAKE2b-256 | baaddf2fe4bed8762c013f3cde5f012091fce86046ae240d66cca246b2bbac8e |
Hashes for pymde-0.1.0-cp38-cp38-macosx_10_9_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | b8fb00d7a60280217241da92f4186f93539f5c764936f2e50058b7e14fc0922f |
|
| MD5 | fa74ebd35ccd1fbe449a96d3d3032938 |
|
| BLAKE2b-256 | 1fc680c8b7a8ae731dbe60d58a12c5c2b552e3999319e3dc63c3683622b5e06e |
Hashes for pymde-0.1.0-cp37-cp37m-win_amd64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | e43ff5a84221a2e2011f7f7ffab6bbcf1eb8979a124409cafec9e0fd8435457f |
|
| MD5 | 69190d883d45dd551fe588be9eb904c4 |
|
| BLAKE2b-256 | e1cc8df87c84b76bd720db4c6b6a7e9559c92c8d1b295b16ad1c97919c1908c5 |
Hashes for pymde-0.1.0-cp37-cp37m-win32.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 4969dac1414b311d7c65b4d742437ccfddb9e63cfe7682d5ccb8d6e4472ed783 |
|
| MD5 | 85a1450ea2d3abf9032a373195878e69 |
|
| BLAKE2b-256 | e106030456527b786cab39ff71306acb71cf4b76771826efde85ea8f8d983ecc |
Hashes for pymde-0.1.0-cp37-cp37m-manylinux2010_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 020e9588a7e2ee99b06b46e8b8a335b4c0c8af6d780e72518610ead8cd6ddddb |
|
| MD5 | 7b04bd945f0d82d98642c4a1518d0ae4 |
|
| BLAKE2b-256 | 8f6f04508b045e659f7af264fa8f2ac5f6c5e38ae2f13c2e46c8e3b2f0f21bf7 |
Hashes for pymde-0.1.0-cp37-cp37m-manylinux2010_i686.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | c4d3369d8f20e02f724d39c70383423bd20f250c4f0be5558d7aa0c438c01846 |
|
| MD5 | 0f9f59a0a52751d45f86ecd77253b957 |
|
| BLAKE2b-256 | da0d47f61fb41a37ae675c20ac5eba0f03889b592cfe491436761a13414f3d4e |
Hashes for pymde-0.1.0-cp37-cp37m-manylinux1_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 2800e47bd3469bfe6166714b0233c294c3f6fdcd6899d68ceac085ebf497feee |
|
| MD5 | f27b72e4edc1de6b258e6d2d627fae8b |
|
| BLAKE2b-256 | c73bb7a899d574729c21bdfdb9a1ad879e6a667e1ced7b29475b3e3ffe7f78ea |
Hashes for pymde-0.1.0-cp37-cp37m-manylinux1_i686.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | a8e26a6bf6679c568ca2b53fc871d3188f0d425771c71334c7011f08ae94abf5 |
|
| MD5 | 413de983a19c618422afd57a8837a544 |
|
| BLAKE2b-256 | 5f7e046fe8e7948338d4f7f85f3e98fe24901b039ec70ae428676b215f03359f |
Hashes for pymde-0.1.0-cp37-cp37m-macosx_10_9_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 2197918641a24af2d5a79c9b2be0f2ceb16c9766ef229fd9cbf547e68fecdb2e |
|
| MD5 | 7d249d36671a1c30b9686ed41af769c9 |
|
| BLAKE2b-256 | dbe3887cc3fcbd9c4865dab9648718611cf5020478858128315e48beba535ef6 |