A simple way to visualize features on human and other species chromosome ideograms, folk of jordanlab/tagore
Project description
quyuan
quyuan is a folk of tagore with several modifications to make it more suitable for my own use.
Installation
quyuan is a simple Python script with several dependencies.
$ pip install git+https://github.com/tcztzy/quyuan.git
$ quyuan --version
quyuan, version 1.1.2
Requirements
Quick start
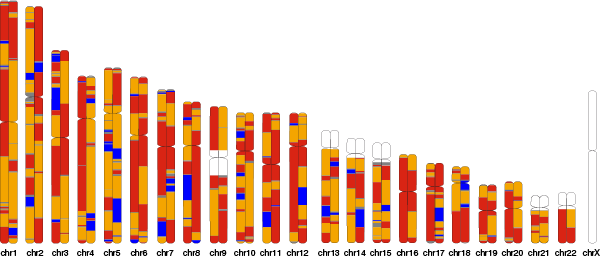
The demo data consists of Catalogue of Somatic Mutations in Cancer (COSMIC) Cancer Gene Census genes and 100 randomly simulated mutations. Points represent single nucleotide variants (i.e. variant present in <3 samples); triangles represent single nucleotide polymorphisms (i.e. variants found in many samples); and short lines (single chromosome) represent known INDEL sites.
$ quyuan --input example_ideogram/test.bed --prefix example_ideogram/example -vf
Usage
Usage: quyuan [OPTIONS]
quyuan: a utility for illustrating human chromosomes
https://github.com/tcztzy/quyuan
Options:
--version Show the version and exit.
-i, --input <input.bed> Input BED-like file [required]
-p, --prefix [output file prefix]
Output prefix [Default: "out"]
-b, --build [hg37|hg38|irgsp1] Human genome build to use [Default: hg38]
-f, --force Overwrite output files if they exist already
-ofmt, --oformat [png|pdf|ps|svg]
Output format for conversion
-v, --verbose Display verbose output
--help Show this message and exit.
The input file is a bed-like format, described below. If an output prefix is not specified, the scripts uses "out" as the default prefix.
Helper scripts for converting RFMix and ADMIXTURE outputs are included in the scripts/ folder.
A more complete example of a full chromosome painting using an RFMix output can be seen by running:
rfmix2tagore --chr1 example_ideogram/1KGP-MXL104_chr1.bed \
--chr2 example_ideogram/1KGP-MXL104_chr2.bed \
--out example_ideogram/1KGP-MXL104_tagore.bed
quyuan --input example_ideogram/1KGP-MXL104_tagore.bed \
--prefix example_ideogram/1KGP-MXL104 \
--build hg37 \
--verbose
Input file description
#chr start stop feature size color chrCopy
chr1 10000000 20000000 0 1 #FF0000 1
chr2 20000000 30000000 0 1 #FF0000 2
chr2 40000000 50000000 0 0.5 #FF0000 1
Each column is explained below:
- chr - The chromosome on which a feature has to be drawn
- start - Start position (in bp) for feature
- stop - Stop position (in bp) for feature
- feature - The shape of the feature to be drawn
- 0 will draw a rectangle
- 1 will draw a circle
- 2 will draw a triangle pointing to the genomic location
- 3 will draw a line at that genomic location
- size - The horizontal size of the feature. Should range between 0 and 1.
- color - Specify the color of the genomic feature with a hex value (#FF0000 for red, etc.)
- chrCopy - Specify the chromosome copy on which the feature should be drawn (1 or 2). To draw the same feature on both chromosomes, you must specify the feature twice
Etymology
Qu Yuan (屈原) was a Chinese patriot poet and politician living in c.340BC - 278BC.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.