Simulate the continuous-time threshold model on static networks.
Project description
Simulate the continuous-time threshold model on static networks using Gillespie’s SSA. The networks can be directed and weighted.
Install
git clone https://github.com/benmaier/thresholdmodel.git
pip install ./thresholdmodelExample
Simulate on an ER random graph.
import numpy as np
import networkx as nx
import matplotlib.pyplot as plt
from thresholdmodel import ThreshModel
N = 1000
k = 10
thresholds = 0.1
initially_activated = np.arange(100)
G = nx.fast_gnp_random_graph(N, k/(N-1.0))
Thresh = ThreshModel(G,initially_activated,thresholds)
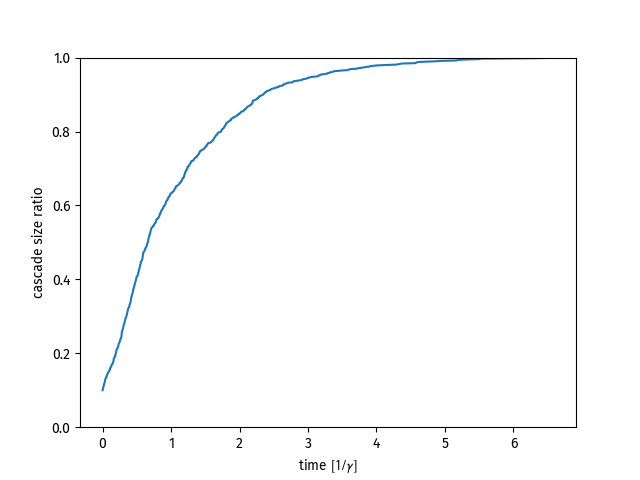
t, a = Thresh.simulate()
plt.plot(t,a)
plt.show()API
Simulate
Given a networkx-Graph object G (can be a networkx.DiGraph, too), and values for initially_activated and thresholds, simulate like this
Thresh = ThreshModel(G,initially_activated,thresholds)
t, a = Thresh.simulate()t is a numpy.ndarray containing the times at which node activations happened. a is a numpy.ndarray containing the relative cascade size at the corresponding time in t. Note that the whole process is modeled as a Poisson process such that the time t will be given in units of the node activation rate gamma = 1.0. If you want to simulate for another node activation rate, simply rescale time as t /= gamma.
When the simulation is started with the save_activated_nodes=True flag, a list of activated nodes per time leap is saved in ThreshModel.activated_nodes.
t, a = Thresh.simulate(save_activated_nodes=True)
print(Thresh.activated_nodes)You can repeat a simulation with the same initial conditions by simply calling Thresh.simulate() again, all the necessary things will be reset automatically.
Set initially activated nodes
Set nodes 3, 5, and 8 to be activated initially.
initially_activated = [3, 5, 8] # this could also be a numpy arrayChoose 20% of all nodes randomly to be activated initially. When the simulation is restarted, the same nodes will be chosen as initial conditions.
initially_activated = 0.2Choose 35 randomly selected nodes to be activated initially. When the simulation is restarted, the same nodes will be chosen as initial conditions.
initially_activated = 35Set thresholds
Activation thresholds can be set for all nodes
thresholds = np.random.rand(G.number_of_nodes())Note that thresholds need to lie in the domain [0,1].
You can also set a universal threshold
thresholds = 0.1Here, 10% of a node’s neighbor’s need to be activated in order for the node to become active, too.
Directed networks
A node will become active if the sufficient number of nodes pointing towards the node are active. This means that the in-degree will be the important measure to determine wether a node will become active.
Weighted networks
If you want to simulate on a weighted network, provide the weight keyword
Thresh = ThreshModel(G,initially_activated,thresholds,weight='weight')Similar to the networkx-documentation: ``weight`` (string, optional (default=``None``)) - The attribute name to obtain the edge weights. E.g.: G.edges[0,1]['weight'].
A focal node will become active when the cumulative edge weights of all activated nodes pointing towards the focal node will reach > threshold*in_degree.
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.