vintools: Michael Vinyard's utilities.
Project description
Data and Dataset API
To load the LARRY dataset, use the following command:
adata = sdq.data.load_LARRY()
To load the EMT simulation dataset, run the following command:
adata = sdq.data.load_EMT_simultion()
Currently, there are four datasets we are currently interested in. The first dataset (1) is a simple, one-attractor, hyperparabola model defined by a set of two ordinary differential equations. The second dataset (2) is a simulation based on the biological phenomena, epithelial to mesenchymal transition (EMT), which has been studied for decades and accumulated a lot of empirical evidence such that simple to complex simulations have been created. The third dataset (3) is a real scRNA-seq dataset collected from a cell-line based model of EMT. The fourth dataset (4) is what will serve as a benchmark for this method; the lineages are traced using lentiviral barcodes, thus we know the ground truth for lineage prediction models.
Dream scenario: It would be interesting to move to an original dataset. There's some work being done using mitochondrial DNA mutations to discover lineages. Perhaps we could work with a collaborating lab to obtain a simple dataset using this technology. This would also enable us to dabble into scATAC.
(1) 2-D one-attractor dataset
There are several variations of this toy dataset, depending on how it is simulated. Thus, it is probably easiest for now to simulate it on the fly since it only takes a few seconds. The easiest way to do this is to install my package, nodescape and run the simulation commands as follows:
def state_eqn(state, t):
x, y = state[0], state[1]
xdot = -x + y ** 2
ydot = -y - x * y
return np.array([xdot, ydot])
Simulate this system of ODEs using:
adata = n.sim.trajectories(
state_eqn, ic_type="meshgrid", number_time_samples=50, time_span=2,
)
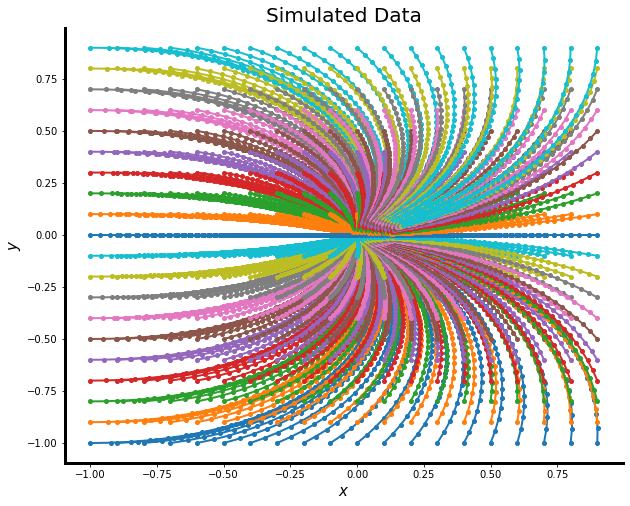
This creates an AnnData object and produces an image in two dimensions. This function is written such that any system of equations defined in state_eqn above can be simulated using an ODE integrator.
AnnData object with n_obs × n_vars = 20000 × 2
obs: 'time', 'trajectory', 'timepoint'
uns: 'number_of_timepoints', 'number_of_trajectories'
Here's a plot of the simulated data corresponding to the command above with the given parameters:
Saving the dataset is easy, jus do the following:
adata.write("/path/to/adata.h5ad")
(2) Simulated EMT dataset
For the EMT dataset, I put in some work to simulate this and there are many paramters. The rationale for how parameters, initial conditions, etc. were chosen are summarized in this notebook. I've adapted a data loader function specifically for this dataset, which can be called using the following.
To load the simulated EMT dataset:
import nodescape as n
adata = n.ut.load_EMT_simulation()
There are no required parameters, but if you want to save it locally (downloading to memory each time only takes a few seconds, ~120MB), you can use the parameter save_local="/path/to/adata.h5ad". Either way, this presents you wtih the AnnData object for the EMT dataset.
AnnData object with n_obs × n_vars = 1080000 × 4
obs: 'time', 'i2', 'trajectory', 'training', 'validation', 'test'
uns: 'number_of_trajectories'
obsm: 'X_pca'
Here's a 2-D PCA plot of the data colored by time, which can be obtained by calling n.ut.pca(adata, plot=True):
This dataset is really dense, in terms of time increments. In fact, I have been thinning it out to get it to work with neural ODEs. I use the following command, which downsamples evenly over time. Then I split it into test/train/validtion (all contained in the data class).
adata = n.ut.downsample(adata, 0.05)
data = n.ml.split_test_train(adata)
I will work on the remaining two datasets . . .
(3) Real EMT dataset (McFaline et al., 2018)
To load the real EMT dataset:
(4) LARRY dataset (Weinreb et al., 2020)
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.