Assess the quality of metagenome-assembled viral genomes.
Project description
CheckV is a fully automated command-line pipeline for assessing the quality of metagenome-assembled viral genomes, including identification of host contamination for integrated proviruses, estimating completeness for genome fragments, and identification of closed genomes.
The pipeline can be broken down into 4 main steps:
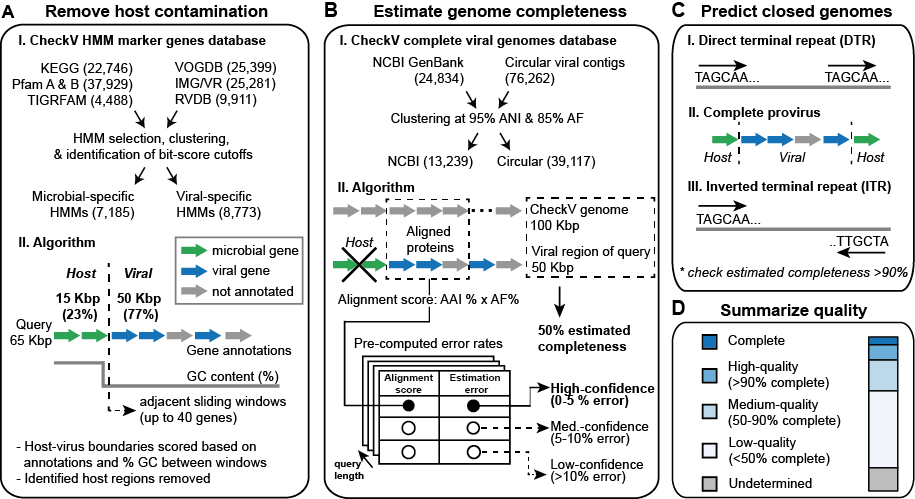
A: Remove host contamination. CheckV identifies and removes non-viral regions on proviruses. Genes are first annotated based on comparison to a custom database of HMMs that are highly specific to either viral or microbial proteins. Next, the program compares the gene annotations and GC content between a pair of sliding windows that each contain up to 40 genes. This information is used to compute a score at each intergenic position and identify host-virus boundaries.
B: Estimate genome completeness. CheckV estimates genome completeness in two stages. First, proteins are compared to the CheckV genome database using AAI (average amino acid identity), completeness is computed as a simple ratio between the contig length (or viral region length for proviruses) and the length of matched reference genomes, and a confidence level is reported. In some cases, a contig won't have a high- or medium-confidence estimate based on AAI. In these cases, a more sensitive but less accurate approach is used based on HMMs shared between the contig and CheckV reference genomes (ANI: average nucleotide identity; AF: alignment fraction)
C: Predict closed genomes. Closed genomes are identified based either on direct terminal repeats (DTRs; often indicating a circular sequence), flanking virus-host boundaries (indicating a complete prophage), or inverted terminal repeats (ITRs; believed to facilitate circularization and recombination). Whenever possible, these predictions are validated based on the estimated completeness obtained in B (e.g. completeness >90%). DTRs are the most reliable and most common indicator of complete genomes.
D: Summarize quality. Based on the results of A-C, CheckV generates a report file and assigns query contigs to one of five quality tiers: complete, high-quality (>90% completeness), medium-quality (50-90% completeness), low-quality (<50% completeness), or undetermined quality. These quality tiers are consistent with and expand upon those outlined in the MIUViG standards.
Installation
There are two methods to install CheckV:
- Using
conda:
conda install -c conda-forge -c bioconda checkv
- Using
pip:
pip install checkv
If you decide to install CheckV via pip, make sure you also have the following external dependencies installed:
- BLAST+ (v2.5.0)
- DIAMOND (v0.9.30)
- HMMER (v3.3)
- Prodigal (v2.6.3)
The versions listed above were the ones that were properly tested. Different versions may also work.
CheckV database
Whichever method you choose to install CheckV you will need to download the database in order to use it:
checkv download_database ./
Or (to download and extract manually):
wget https://portal.nersc.gov/CheckV/checkv-db-v0.6.tar.gz
tar -zxvf checkv-db-v0.6.tar.gz
And update your environment (optional):
export CHECKVDB=/path/to/checkv-db
Some users may wish to update the database using their own complete genomes (available in next release):
checkv update_database /path/to/checkv-db /path/to/updated-checkv-db genomes.fna
Quick start
There are two ways to run CheckV:
- Using individual commands for each step in the pipeline in the following order:
checkv contamination input_file.fna output_directory -t 16
checkv completeness input_file.fna output_directory -t 16
checkv repeats input_file.fna output_directory
checkv quality_summary input_file.fna output_directory
- Using a single command to run the full pipeline (available in next release):
checkv end_to_end input_file.fna output_directory -t 16
- For a full listing of checkv programs and options, use:
checkv -handcheckv <program> -h
Automated testing
CheckV uses the pytest framework for checking the accuracy of the outputs of its main functions. The testing pipeline executes the contamination, completeness, repeats and quality_summary modules sequentially and then compares the output files with the ground truth.
To avoid introducing unwanted breaking changes, we recommend developers and users to execute the tests before committing any changes or submitting new PRs to the repository.
pytest --database=/path/to/checkv-db --threads=16
Known Issues
- Bug in completeness module (v0.5.1) that suppresses output of HMM-based results
- DIAMOND v0.9.30 does not work on OSX
Output files
quality_summary.tsv
This contains integrated results from the three main modules and should be the main output referred to. Below is an example to demonstrate the type of results you can expect in your data:
| contig_id | contig_length | genome_copies | gene_count | viral_genes | host_genes | checkv_quality | miuvig_quality | completeness | completeness_method | contamination | prophage | termini |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 23404 | 1.0 | 48 | 0 | 0 | Not-determined | Genome-fragment | NA | NA | 0 | No | NA |
| 2 | 9837 | 1.0 | 14 | 4 | 2 | Low-quality | Genome-fragment | 10.73 | AAI-based | 41.92 | Yes | NA |
| 3 | 49521 | 1.0 | 68 | 26 | 1 | Medium-quality | Genome-fragment | 80 | HMM-based | 0 | No | NA |
| 4 | 81346 | 2.1 | 104 | 3 | 12 | High-quality | High-quality | 100 | AAI-based | 0 | No | NA |
| 5 | 55870 | 1.02 | 66 | 11 | 1 | Complete | High-quality | 100 | AAI-based | 0 | No | 33-bp-DTR |
| 6 | 53208 | 1.0 | 80 | 29 | 7 | Complete | High-quality | 100 | AAI-based | 9.79 | Yes | complete-prophage |
In the example, above there are results for 6 viral contigs:
- The first 23 kb contig has no completeness prediction, which is indicated by 'Not-determined' for the 'checkv_quality' field. This contig also has no viral genes identified, so there's a chance it may not even be a virus.
- The second 9.8 kb contig is classified as 'Low-quality' since its completeness is <50%. This is estimate is based on the 'AAI' method. Note that only either high- or medium-confidence estimates are reported in the quality_summary.tsv file. You can see 'completeness.tsv' for more details.
- The third contig is considered 'Medium-quality' since its completeness is estimated to be 80%, which is based on the 'HMM' method. This means that it was too novel to estimate completeness based on AAI, but shared an HMM with CheckV reference genomes. Note that the HMM-based method may underestimate the true completeness.
- The fourth contig is classified as High-quality based on a completness of >90%. However, note that value of 'genome_copies' is 2.10. This indicates that the viral genome is represented multiple times in the contig. These cases are quite rare, but something to watch out for.
- The fifth contig is classified as Complete based on the presence of a 33-bp direct terminal repeat and has 100% completeness based on the AAI method. This sequence can condifently treated as a complete genome.
- The sixth contig is classified as Complete based on the presence of multiple host-virus boundaries and an estimated completeness of >90%. Complete prophages and ITRs often have estimated completeness <90%, so greater caution is needed for analying these sequences.
completeness.tsv
A detailed overview of how completeness was estimated:
| contig_id | contig_length | viral_length | aai_expected_length | aai_completeness | aai_confidence | aai_error | aai_num_hits | aai_top_hit | aai_id | aai_af | hmm_completeness | hmm_name | hmm_ref_genomes | hmm_avg_length | hmm_cv_length |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 9837 | 5713 | 53242.8 | 10.7 | high | 3.7 | 10 | DTR_517157 | 78.5 | 34.6 | 5 | 3300001592@Draft_10000123@Draft_1000012363 | 164 | 44645 | 0.152 |
| 2 | 39498 | 39498 | 37309 | 105.8 | medium | 7.7 | 11 | DTR_357456 | 45.18 | 30.46 | 75 | 3300001729@JGI24651J20071_1000011@JGI24651J20071_100001152 | 277 | 40578.1 | 0.145 |
| 3 | 49521 | 49521 | 44960.1 | 110.1 | low | 10.2 | 17 | DTR_091230 | 39.74 | 19.54 | 80 | VOG00187 | 496 | 48703.6 | 0.117 |
| 4 | 23404 | NA | NA | NA | NA | NA | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
In the example, above there are results for 4 viral contigs:
- The first viral contig has an estimated completeness of 10.7% based on the AA-based method. The confidence for this estimate is high, based on a relative estimated error of 3.7%, which is in turn based on the aai_id (average amino acid identity) and aai_af (alignment fraction of contig) to the CheckV reference DTR_517517
- The second contig is considered high-quality with a completeness of 105%. The completeness is >100% because the contig length is a bit longer than the expected genome length of 37,309 bp.
- The third contig is estimated to be 80% complete based on the HMM approach. We do not trust the AAI-based estimate in this case because the confidence level is low and the aai_error is >10%. The viral HMM used for completeness estimation is VOG00187
- The last contig doesn't have any hits based on AAI and doesn't have any viral HMMs, so there's nothing we can do with this sequence
contamination.tsv
A detailed overview of how contamination was estimated:
| contig_id | contig_length | viral_length | host_length | total_genes | viral_genes | host_genes | region_types | region_lengths | region_coords | region_genes |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 23404 | 0 | 0 | 48 | 0 | 0 | unclassified | 23404 | 1-23404 | 1-48 |
| 2 | 13497 | 13497 | 0 | 9 | 1 | 0 | viral | 497 | 1-13497 | 1-9 |
| 3 | 9837 | 5713 | 4124 | 14 | 4 | 2 | host,viral | 4124,5713 | 1-4124,4125-9837 | 1-6,7-14 |
| 4 | 81346 | 0 | 81346 | 104 | 3 | 12 | host | 81346 | 1-81346 | 1-104 |
In the example, above there are results for 4 viral contigs:
- The first viral contig doesn't have any identified viral or host genes, so it's labeled unclassified. Reported as: Prophage='No', Contamination='No' in quality_summary.tsv
- The second viral contig appears to be free of contamination. Reported as: Prophage='No', Contamination='No' in quality_summary.tsv
- The third 9.8 kb contig is identified as a provirus based on host genes identified on the left portion of the contig (bases 1 to 4124 and genes 1-6). Reported as: Prophage='Yes', Contamination=30.5% in quality_summary.tsv
- The fourth 81 kb contig has 104 genes, including 3 viral and 12 host genes. No virus-host boundaries were identified. The overall sequence is classified as host (host genes > viral genes), though CheckV was not designed to discriminate viruses from non-viral sequences. Reported as: Prophage='No', Contamination=0% in quality_summary.tsv
repeats.tsv
A detailed overview of terminal repeats and genome copy number estimation:
| contig_id | contig_length | genome_copies | repeat_type | repeat_length | repeat_count | repeat_dust_length | repeat_flagged | reason |
|---|---|---|---|---|---|---|---|---|
| 1 | 55870 | 1.02 | DTR | 33 | 2 | 0 | No | NA |
| 2 | 35087 | 1 | ITR | 55 | 1 | 0 | No | NA |
| 3 | 14082 | 1 | DTR | 20 | 11 | 0 | Yes | repetetive |
| 4 | 143514 | 1.03 | DTR | 45 | 2 | 40 | Yes | low-complexity |
| 5 | 336658 | 2.10 | NA | NA | NA | NA | NA | NA |
In the example, above there are results for 5 viral contigs:
- The first viral contig has a direct terminal repeat of 33 bp
- The second viral contig has an inverted direct terminal repeat of 55 bp
- The third viral contig has DTR of 20 bp. However the 20-bp repeat occurs 11 times on the 14 kb contig, suggesting it is a repetetive element rather than a complete genome. This contig is flagged and will not appear in the quality_summary.tsv output
- The fourth viral contig has DTR of 45 bp. However 40/45-bp are classified as low complexity by dustmasker (e.g. AAAAA...). This will not appear in the quality_summary.tsv output
- The fifth viral contig has a genome_copies value of 2.10. This is calcuated by counting 1 kb sequences across the contig. This value suggests the contig represents two of the exact same viral genomes. Though rare, it's thought that these can occur due to assembly errors.
Frequently asked questions
Q: What is the difference between AAI- and HMM-based completeness?
A: AAI-based completeness was designed to be very accurate and can be trusted when the confidence is medium or high. HMM-based completeness was designed to confidently estimate the minimum completeness. So a value of 50% indicates that we can be 95% sure that the viral contig is at least 50% complete. But it may be more complete, so this should be taken into consideration when analyzing CheckV output.
Q: What is the meaning of the genome_copies field?
A: This is a measure of how many times the viral genome is represented in the contig. Most times this is 1.0 (or very close to 1.0). In rare cases assembly errors may occur in which the contig sequence represents multiple concatenated copies of the viral genome. In these cases genome_copies will exceed 1.0.
Q: Why does my DTR contig have <100% estimated completeness?
A: If the estimated completeness is close to 100% (e.g. 90-110%) then the query is likely complete. However sometimes incomplete genome fragments may contain a direct terminal repeat (DTR), in which case we should expect their estimated completeness to be <90%, and sometimes much less. In other cases, the contig will truly be circular, but the estimated completeness is incorrect. This may also happen if the query a complete segment of a multipartite genome (common for RNA viruses). By default, CheckV uses the 90% completeness cutoff for verification, but a user may wish to make their own judgement in these ambiguous cases.
Q: Why is my DTR contig predicted as a provirus?
A: CheckV classifies a sequence as a provirus if it is contains a host region (usually occuring on one just side of the sequence). A DTR sequence represents a complete viral genome, so these predictions are at odds with eachother and indicate either a false positive DTR prediction, or a false positive provirus prediction. By default, CheckV considers these complete genomes, but a user may wish to make their own judgement in these ambiguous cases.
Q: Why is my sequence considered "high-quality" when it has high contamination?
A: CheckV determines sequence quality solely based on completeness. Host contamination is easily removed, so is not factored into these quality tiers.
Q: I performed binning and generated viral MAGs. Can I use CheckV on these?
A: CheckV can estimate completeness but not contamination for these. Additionally, you'll need to concatentate the contigs from each MAG into a single sequence prior to running CheckV.
Q: Can I apply CheckV to eukaryotic viruses?
A: Probably, but this has not been tested. The reference database includes a large number of genomes and HMMs that should match eukaryotic genomes. However, CheckV may report a completeness <90% if your genome is a single segment of a segmented viral genome. CheckV may also classify your sequence as a provirus if it contains a large island of metabolic genes commonly found in bacteria/archaea.
Q: Can I use CheckV to predict (pro)viruses from whole (meta)genomes?
A: Possibly, though this has not been tested.
Q: How should I handle putative "closed genomes" with no completeless estimate?
A: In some cases, you won't be able to verify the completeness of a sequence with terminal repeats or provirus integration sites. DTRs are a fairly reliable indicator (>90% of the time) and can likely be trusted with no completeness estimate. However, complete proviruses and ITRs are much less reliable indicators, and therefore require >90% estimated completeness.
Q: Why is my contig classified as "undetermined quality"?
A: This happens when the sequence doesn't match any CheckV reference genome with high enough similarity to confidently estimate completeness and doesn't have any viral HMMs. There are a few explanations for this, in order of likely frequency: 1) your contig is very short, and by chance it does not share any genes with a CheckV reference, 2) your contig is from a very novel virus that is distantly related to all genomes in the CheckV database, 3) your contig is not a virus at all and so doesn't match any of the references.
Q: How should I handle sequences with "undetermined quality"?
A: While it is not possible to estimate completeness for these, you may choose to still analyze sequences above a certain length (e.g. >30 kb).
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.