Compositional Perturbation Autoencoder (CPA)
Project description
CPA - Compositional Perturbation Autoencoder
What is CPA?
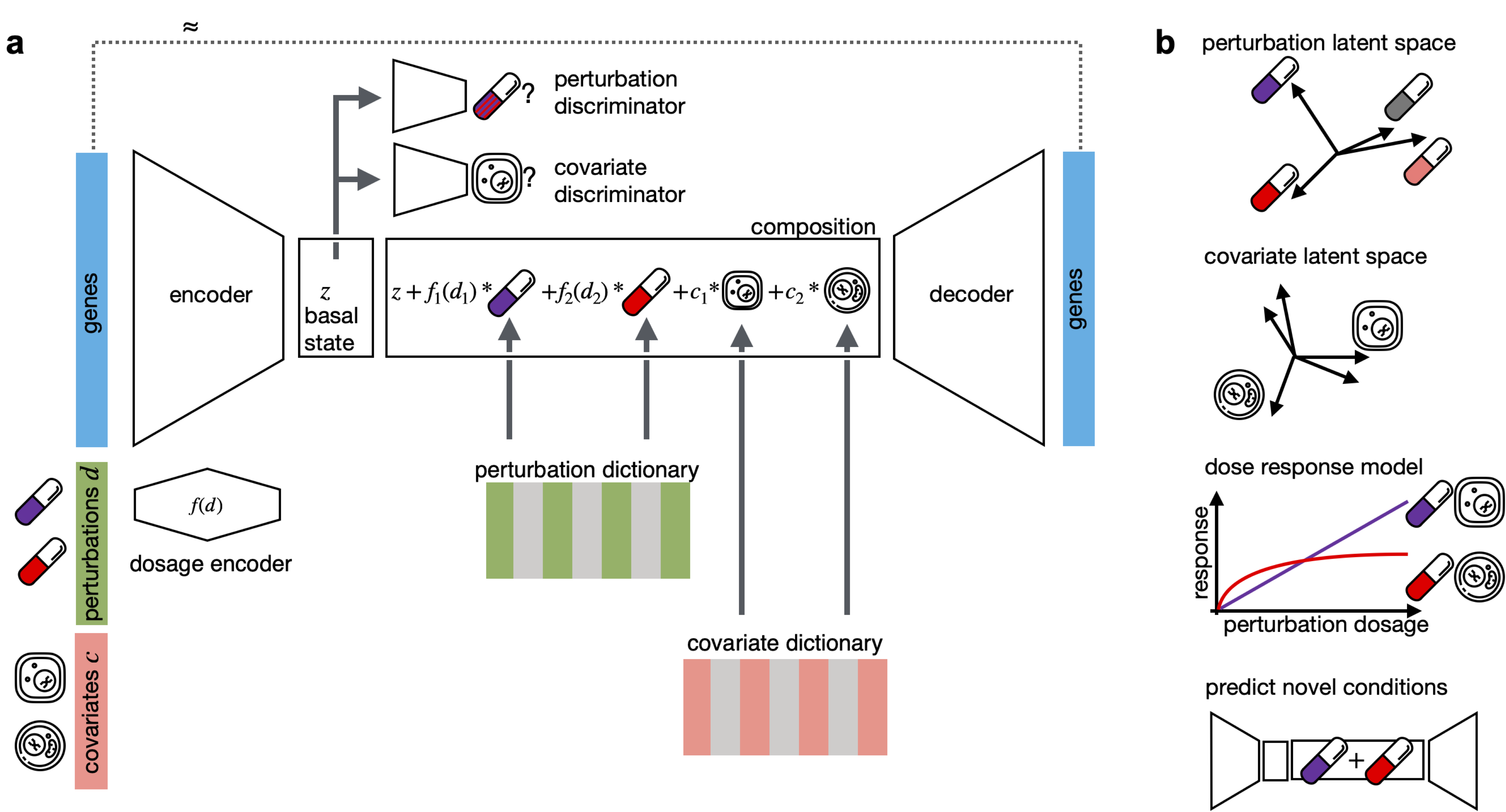
CPA is a framework to learn effects of perturbations at the single-cell level. CPA encodes and learns phenotypic drug response across different cell types, doses and drug combinations. CPA allows:
- Out-of-distribution predictions of unseen drug combinations at various doses and among different cell types.
- Learn interpretable drug and cell type latent spaces.
- Estimate dose response curve for each perturbation and their combinations.
- Access the uncertainty of the estimations of the model.
Usage and installation
See here for documentation and tutorials.
Support and contribute
If you have a question or new architecture or a model that could be integrated into our pipeline, you can post an issue
Acknowledgment
This code is inspired by an early implementatiom by Pierre Boyeau using scvi-tools.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
cpa-tools-0.2.11.tar.gz
(35.7 kB
view hashes)
Built Distribution
cpa_tools-0.2.11-py3-none-any.whl
(36.6 kB
view hashes)
Close
Hashes for cpa_tools-0.2.11-py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 06a9f08debdc15a3c88ccd98a23c7692b4bc51d08ed21bab17ca0af127b8ff6c |
|
| MD5 | 9a2cd1e259f32923214ec2572a52e56f |
|
| BLAKE2b-256 | e95e97b6070010f13949a7488dd1cc50fa0b3fc9294cebd4d6db3dca7c9e171d |