Compositional Perturbation Autoencoder (CPA)
Project description
CPA - Compositional Perturbation Autoencoder 


What is CPA?
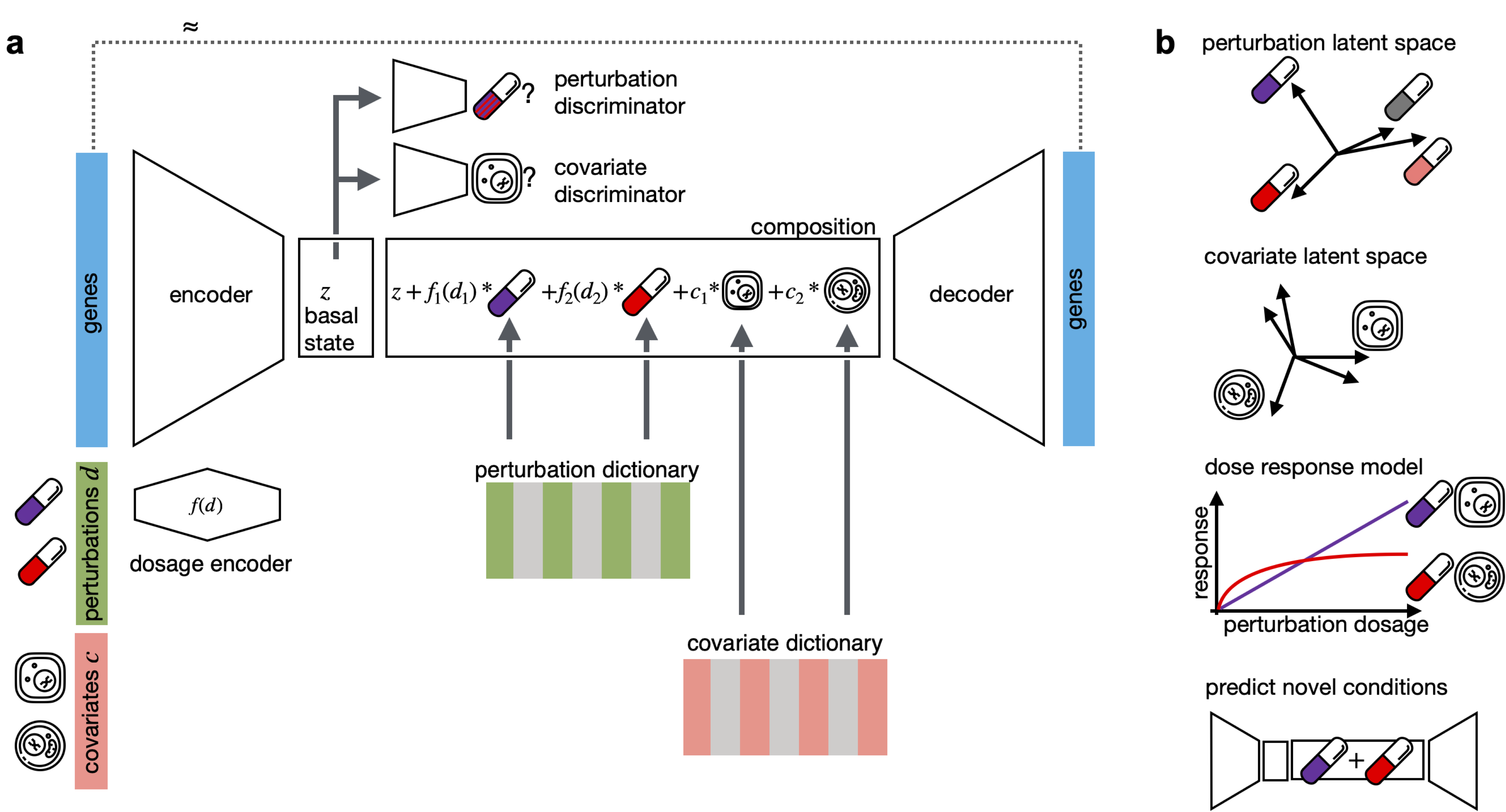
CPA is a framework to learn the effects of perturbations at the single-cell level. CPA encodes and learns phenotypic drug responses across different cell types, doses, and combinations. CPA allows:
- Out-of-distribution predictions of unseen drug and gene combinations at various doses and among different cell types.
- Learn interpretable drug and cell-type latent spaces.
- Estimate the dose-response curve for each perturbation and their combinations.
- Transfer pertubration effects from on cell-type to an unseen cell-type.
Installation
Requirements
Conda Environment
We recommend using Anaconda/Miniconda to create a conda environment for using CPA. You can create a python environment using the following command:
conda create -n cpa python=3.8
Then, you can activate the environment using:
conda activate cpa
Pytorch
CPA is implemented in Pytorch and requires Pytorch version >= 1.13.1.
OSX
pip install torch==1.13.1
Linux and Windows
If you have access to GPUs, you can install the GPU version of Pytorch following the instructions here.
Sample command for installing Pytorch 1.13.1 on different CUDA versions:
# ROCM 5.2 (Linux only)
pip install torch==1.13.1+rocm5.2 --extra-index-url https://download.pytorch.org/whl/rocm5.2
# CUDA 11.6
pip install torch==1.13.1+cu116 --extra-index-url https://download.pytorch.org/whl/cu116
# CUDA 11.7
pip install torch==1.13.1+cu117 --extra-index-url https://download.pytorch.org/whl/cu117
# CPU only
pip install torch==1.13.1+cpu --extra-index-url https://download.pytorch.org/whl/cpu
Installing CPA
You can install CPA using pip:
pip install cpa-tools
How to use CPA
Several tutorials are available here to get you started with CPA. The following table contains the list of tutorials:
How to optmize CPA hyperparamters for your data
Datasets and Pre-trained models
Datasets and pre-trained models are available here.
Support and contribute
If you have a question or new architecture or a model that could be integrated into our pipeline, you can post an issue
Reference
If CPA is helpful in your research, please consider citing the Lotfollahi et al. 2023
@article{lotfollahi2023predicting,
title={Predicting cellular responses to complex perturbations in high-throughput screens},
author={Lotfollahi, Mohammad and Klimovskaia Susmelj, Anna and De Donno, Carlo and Hetzel, Leon and Ji, Yuge and Ibarra, Ignacio L and Srivatsan, Sanjay R and Naghipourfar, Mohsen and Daza, Riza M and
Martin, Beth and others},
journal={Molecular Systems Biology},
pages={e11517},
year={2023}
}
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distributions
Built Distribution
Hashes for cpa_tools-0.8.0-py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 836e922aef3b1e1a475c3024562779eeab41a6af6db150218875205d362dcea2 |
|
| MD5 | 359e9552c584c6a5ca0405ec085f91c5 |
|
| BLAKE2b-256 | 1679f803c0791689c7cc363e917e64a66fcb9186d820ebdec5ffd9ceac0cb4f6 |