Spectral Clustering
Project description
Spectral Clustering
Note
We are currently adding new functionalities to this library to include some algorithms to appear in an upcoming paper. We are updating the APIs as well.
If you depend on our old API, please use an older version of this library:
pip3 install spectralcluster==0.1.0
Overview
This is a Python re-implementation of the spectral clustering algorithm in the paper Speaker Diarization with LSTM.
Disclaimer
This is not a Google product.
This is not the original C++ implementation used by the paper.
Dependencies
- numpy
- scipy
- scikit-learn
Installation
Install the package by:
pip3 install spectralcluster
or
python3 -m pip install spectralcluster
Tutorial
Simply use the predict() method of class SpectralClusterer to perform
spectral clustering. The example below should be closest to the original C++
implemention used my our ICASSP 2018 paper.
from spectralcluster import configs
labels = configs.icassp2018_clusterer.predict(X)
The input X is a numpy array of shape (n_samples, n_features),
and the returned labels is a numpy array of shape (n_samples,).
You can also create your own clusterer like this:
from spectralcluster import SpectralClusterer
clusterer = SpectralClusterer(
min_clusters=2,
max_clusters=7,
autotune=None,
laplacian_type=None,
refinement_options=None,
custom_dist="cosine")
labels = clusterer.predict(X)
For the complete list of parameters of SpectralClusterer, see
spectralcluster/spectral_clusterer.py.
Advanced features
Refinement operations
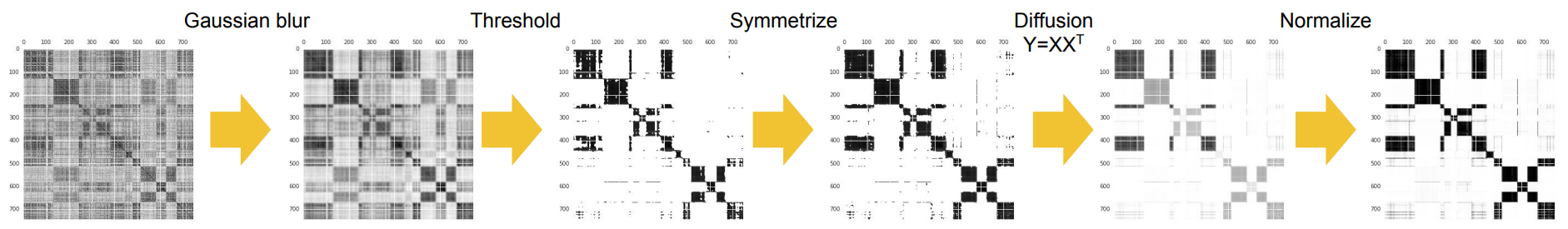
In our ICASSP 2018 paper, we apply a sequence of refinment operations on the affinity matrix, which is critical to the performance on the speaker diarization results.
You can specify your refinment operations like this:
from spectralcluster import RefinementOptions
from spectralcluster import ThresholdType
from spectralcluster import ICASSP2018_REFINEMENT_SEQUENCE
refinement_options = RefinementOptions(
gaussian_blur_sigma=1,
p_percentile=0.95,
thresholding_soft_multiplier=0.01,
thresholding_type=ThresholdType.RowMax,
refinement_sequence=ICASSP2018_REFINEMENT_SEQUENCE)
Then you can pass the refinement_options as an argument when initializing your
SpectralClusterer object.
For the complete list of RefinementOptions, see
spectralcluster/refinement.py.
Laplacian matrix
In our ICASSP 2018 paper,
we apply a refinement operation CropDiagonal on the affinity matrix, which replaces each diagonal element of the affinity matrix by the max non-diagonal value of the row. After this operation, the matrix has similar properties to a standard Laplacian matrix, and it is also less sensitive (thus more robust) to the Gaussian blur operation than a standard Laplacian matrix.
In the new version of this library, we support different types of Laplacian matrix now, including:
- None Laplacian (affinity matrix):
W - Unnormalized Laplacian:
L = D - W - Graph cut Laplacian:
L' = D^{-1/2} * L * D^{-1/2} - Random walk Laplacian:
L' = D^{-1} * L
You can specify the Laplacian matrix type with the laplacian_type argument of the SpectralClusterer class.
Note: Refinement operations are applied to the affinity matrix before computing the Laplacian matrix.
Distance for K-Means
In our ICASSP 2018 paper, the K-Means is based on Cosine distance.
You can set custom_dist="cosine" when initializing your SpectralClusterer object.
You can also use other distances supported by scipy.spatial.distance, such as "euclidean" or "mahalanobis".
Auto-tune
We also support auto-tuning the p_percentile parameter of the RowWiseThreshold refinement operation, which was original proposed in this paper.
You can enable this by passing in an AutoTune object to the autotune argument when initializing your SpectralClusterer object.
Example:
from spectralcluster import AutoTune
autotune = AutoTune(
p_percentile_min=0.60,
p_percentile_max=0.95,
init_search_step=0.01,
search_level=3)
For the complete list of parameters of AutoTune, see
spectralcluster/autotune.py.
Constrained spectral clustering
We also implemented 2 constrained spectral clustering methods:
If you pass in a ConstraintOptions object when initializing your SpectralClusterer object, you can call the predict function with a constraint_matrix.
Example usage:
from spectralcluster import constraint
ConstraintName = constraint.ConstraintName
constraint_options = constraint.ConstraintOptions(
constraint_name=ConstraintName.ConstraintPropagation,
apply_before_refinement=True,
constraint_propagation_alpha=0.6)
clusterer = spectral_clusterer.SpectralClusterer(
max_clusters=2,
refinement_options=refinement_options,
constraint_options=constraint_options,
laplacian_type=LaplacianType.GraphCut,
row_wise_renorm=True)
labels = clusterer.predict(matrix, constraint_matrix)
The constraint matrix can be constructed from a speaker_turn_scores list:
from spectralcluster import constraint
constraint_matrix = constraint.ConstraintMatrix(
spk_turn_entries, threshold=1).compute_diagonals()
Citations
Our paper is cited as:
@inproceedings{wang2018speaker,
title={Speaker diarization with lstm},
author={Wang, Quan and Downey, Carlton and Wan, Li and Mansfield, Philip Andrew and Moreno, Ignacio Lopz},
booktitle={2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP)},
pages={5239--5243},
year={2018},
organization={IEEE}
}
Misc
Our new speaker diarization systems are now fully supervised, powered by uis-rnn. Check this Google AI Blog.
To learn more about speaker diarization, here is a curated list of resources: awesome-diarization.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Hashes for spectralcluster-0.2.2-py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 1564f78ff2a96fadcc2f100cbf707224a664aa5938fd2fd6ae82495ffc15e1e6 |
|
| MD5 | 37d59dc4aa3dba608afe5f8095c5c3eb |
|
| BLAKE2b-256 | 0b40acf0b3196e48485f1321f890aa276854669aa2db27e96fcb8a50bf02f754 |